NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The adaptive immune system of jawed vertebrates is based on a vast, anticipatory repertoire of specific antigen receptors, immunoglobulins (Ig) in B-lymphocytes and T-cell receptors (TCR) in T-lymphocytes. The Ig and TCR diversity is generated by a process called V(D)J recombination, which is initiated by the RAG recombinase. Although RAG activity is very well conserved, the regulated accessibility of the antigen receptor genes to RAG has evolved with the species' organizational structure, which differs most significantly between fishes and tetrapods. V(D)J recombination was primarily characterized in developing lymphocytes of mice and human beings and is often described as an ordered, two-stage program. Studies in rabbit, chicken and shark show that this process does not have to be ordered, nor does it need to take place in two stages to generate a diverse repertoire and enable the expression of a single species of antigen receptor per cell, a restriction called allelic exclusion.
Introduction
Origins of the Adaptive Immune System
V(D)J recombination is the process by which antigen receptors, immunoglobulin (Ig) and T-cell receptor (TCR), are assembled for expression during development of the respective B- and T-lymphocytes. Somatic rearrangement of the V (variable), D (diversity) and J (joining) gene segments1 is initiated by the recombinase RAG (recombination-activating gene)2,3 in a cut-and-paste process that entails joining of these separate gene components to encode the V region, the N-terminus of the receptor polypeptide. The V region is 100-120 amino acid long and forms the ligand-binding site in heterodimers of heavy (H) and light (L) chains of Ig, the alpha and beta chains of TCRαβ and the gamma and delta chains of TCRγδ.
RAG and lymphocytes expressing Ig and TCR are present in all jawed vertebrates (Fig. 1), from cartilaginous fishes to mammals. Neither RAG nor the rearranging receptors are found in protochordates or lamprey and hagfish, which suggests that the present RAG function became established in a vertebrate ancestor sometime in the 80 million years between the divergence of jawless fishes and cartilaginous fishes.4,5 Extensive duplication events, either two whole-genome duplications or one genome-wide duplication and multiple segmental duplications occurred before and after divergence of jawless fishes.6-9 The incipience and evolution of the adaptive immune system took place during this period of extensive genomic restructuring.10
The origin of the rearranging genes was first suggested by Sakano and coworkers,11 who remarked that the recognition motifs (recombination signal sequences, RSS) adjacent the V(D)J gene segments were reminiscent of signals found at the termini of integrated transposable elements. Subsequently the chemistry of the RAG-mediated pathway was found to resemble those described for transpositional recombination by mobile elements.12 With the growing availability of genome information from different species, it became feasible to attempt delving into the origins of RAG and its recognition sequences. Terminal inverted repeats with motifs and spacer interval similar to RSS were observed in the Transib transposon in nematodes, insects and sea urchin.13 RAG is composed of two components and both RAG1- and RAG2-like sequences were detected in the sea urchin genome, although their function is not yet clear.14 These discoveries, together with demonstration of latent transposase activity in RAG,15,16 argue for RAG having been part of a DNA transposon that was introduced early into the vertebrate lineage, evolving to its role of V(D)J recombinase by retaining the excision component of transposase activity.17-19 The presence of RAG sequences in echinoderms could indicate entry of the transposon at a far earlier time and lost in certain phyla and classes (protochordates, jawless fishes) but retained in others, or else a separate horizontal transfer in jawed vertebrates.
It is hypothesized that in the ancestral vertebrate the RAG transposon became integrated into a V-like gene, splitting it into two components that can rejoin after RAG-induced double-strand breakage and removal of the intervening DNA.11 The cleavage occurs in the same place due to RSS recognition, but because of the nucleotide loss and/or gain arising from the repair process, the new joints would be varied in sequence (next section). Breakage and repair of DNA induced by RAG thus generates molecular heterogeneity, which may arguably have been the selecting factor if the original V gene had an immune function that was enhanced by diversified sequences.
V(D)J recombination became established in early vertebrates about 500 million years ago and is the process that assembles Ig and TCR genes in all species and the species-specific receptor genes like the IgNAR (new antigen receptor) and NAR-TCR in sharks20,21 and TCRμ in marsupials.22 This chapter deals mainly with comparative studies on the Ig gene system. There is considerably more information on antibody in early vertebrates, due to the much longer history of studies of Ig protein and to the relative ease of detecting VH sequences across species with heterologous probes. The IgM molecule is very well conserved from sharks to mammals23,24 in overall sequence and structure, being the antigen receptor on naive B-cells that in plasma cells is secreted as a polymeric antibody, usually a pentamer. TCR cDNA sequences characterized in all animals show that they are cell surface receptors only. TCRαβ and TCRγδ have been cloned from all classes of jawed vertebrates,25-29 including all three major groups of mammals (marsupials such as opossum,30 monotremes such as duckbill platypus31 and placentals of most orders, including rodents, rabbits, ruminants and primates).
There are two evolutionarily conserved features of V(D)J recombination: the mechanism of RAG action and the regulation of this process to ensure one end result—that only one kind of antigen receptor is expressed per cell (for a review, see ref. 32). This restriction is called allelic exclusion. Although the recombination pathway mediated by RAG is well conserved, the regulated accessibility of the antigen receptor genes to RAG has evolved with the organizational structure, which differs most significantly between cartilaginous fishes and tetrapods (Fig. 2).
V(D)J Rearrangement
RAG Recognition and Joint Resolution
The rearranging elements—the gene segments V, D and J with their adjacent RSS—are present in all classes of jawed vertebrates, as are the key enzymes involved in DNA nicking and modification, RAG1/RAG2 and terminal deoxynucleotidyl transferase (TdT). Although these lymphocyte-specific enzymes have been studied almost entirely in mouse or in vitro systems,33 their highly conserved mode of action in other animals may be deduced. First, pairwise recognition of the RSS is required and the RSS pair to be recombined must consist of one RSS containing a 12-bp spacer and the other a 23-bp spacer (“12/23 rule”).1 In all vertebrates where the genomic organization of the gene segments has been determined, the RSS that flank potentially recombinogenic gene segments reflect this pairing relationship.
RAG initiates the pathway that leads to double-strand breaks at either gene segment and the coding ends being subsequently joined by the cell's DNA repair processes (Fig. 3). Double-strand breakage is obtained in a transesterification reaction that results in a covalently closed hairpin on the coding end and a free blunt RSS at the other. The hairpin is opened asymmetrically, creating an overhang with inverted repeat, some of which is occasionally retained (P region) as part of the ligated joint.
The presence of P region is thus indicative of a hairpin intermediate created during the double-strand break and joining process. Examination of the VD and DJ junctions in Ig H chains and TCR β and δ chains, or VJ junctions in Ig L chains and TCR α and γ chains, the portion of the V sequence called CDR3 (complementarity-determining region 3), shows germline contribution (V and J gene segment flanks, portions of D gene sequence) and occasional P region in all animals, suggesting that V(D)J recombination at different loci and in various species undergo the same unique process involving hairpinned coding ends.
Selection for Junctional Diversification
A second category of somatically-generated additions at the junction is N region, which consists of non-templated, mostly GC-rich sequences catalyzed by TdT34,35 that, together with coding end-processing mediated by exonucleases, are the main contributors to generating the diversification at the junctions of the rejoined gene segments.
The other members of the mammalian Family X DNA polymerases, to which TdT belongs, are not restricted to precursor lymphocytes, but two of them, Pol μ and Pol λ, are also involved at different points during coding end-processing and appear to modulate the extent of coding end nucleolytic processing.36 TdT and Pol μ are very closely related37 and their presence in fishes,38 in contrast to the single-copy ancestral form in the urochordate Ciona (tunicates, Fig. 1), suggests that the lymphocyte-specific TdT evolved to its current role in the immune system by the time of divergence of cartilaginous fishes. The early involvement of TdT in the evolution of V(D)J recombination reflects its importance in amplifying the selected attributes conferred by gene rearrangement: sequence and sequence length variation as a result of RAG-induced breakage.
Diversification mechanisms like mutation or gene conversion may exist in invertebrates39 and predate the rearranging Ig gene system, but these processes do not generate sequence length diversity repeatedly and reliably in one location that will tolerate a loop size spectrum of 2-23 amino acids in the human H chain40 or 9-13 in the shark L chain.41 The greatest contribution to the combining site topology is thus made by the variable CDR3. From crystallographic studies of antigen-antibody complexes H chain CDR3 appears to play the most significant role, not only in the number of contacts with antigen but also in its potential for conformational changes for “induced fit” upon binding ligand.42,43
Novel Rearranging Genes in Sharks and Marsupials
Some species require antigen receptor diversity additional to that provided by heterodimer specificities of the Ig/TCR repertoire. They are (1) shark IgNAR20: neither Ig nor TCR but an early divergent gene, (2) shark NAR-TCR21: TCR isoforms, produced by grafting an additional V region onto an existing TCRδ rearrangement by splicing, forming two successive V regions and (3) marsupial TCRμ22: a hybrid of Ig and TCR components whose product also contains two joined V regions. IgNAR is a secreted serum protein and the other two presumably are active in cell-mediated processes.
IgNAR is a dimer but the V regions are not paired; the ligand-binding site is thus a single V region. The IgNAR V region is generated by four rearrangements—V, three D and J gene segments—providing highly variable and exceptionally plastic CDR3 that are postulated to adopt multiple conformations for induced-fit binding.44 Ig H chain dimers with single-domain V regions (VHH) are also expressed in camels,44 although these gene segments are part of the IgH locus.45 Shark IgNAR and the camel VHH are the result of convergent evolution, as are shark NAR-TCR and opossum TCRμ. NAR-TCR is part of the shark TCRδ locus, whereas TCRμ in opossum are encoded by independent gene clusters.46
TCRμ, like IgNAR, involves rearrangement of 2-3 D elements. Because TCRμ and IgNAR are encoded by a few miniloci, their repertoire is based solely on CDR3 junctional diversity. The use of a single V domain, some with longer CDR3, in cartilaginous fishes and in mammals suggests that there exists some category of antigens that require ligand-binding sites perhaps more flexible than provided by the classical Ig or TCR heterodimer.
One Receptor Per Cell
Both the strength and the weakness of V(D)J recombination is its random nature. An immensely diverse, anticipatory repertoire is generated concomitant with cell and resource wastage. The process cannot ensure that the V becomes joined in-frame with respect to the J (and C region) sequence, so that least two of three rearrangements attempts are nonfunctional. Moreover, randomly-generated specificities also include those that recognize self components and these are eliminated at the immature lymphocyte stage when triggered by a self ligand. Selection for self-tolerance or mounting an immune response is most efficaciously (i.e., specifically) mediated when only one species of receptor is expressed per cell.47 The last phenomenon, known generally as allelic exclusion, results from regulated RAG access to the recombinogenic elements. V(D)J recombination is lineage- and cell stage-specific, meaning that DNA from nonlymphoid cells or from cells of the incorrect developmental stage are not acted upon by RAG.48 There are, however, some interesting exceptions in cartilaginous fishes and these are described in a later section (“Rearrangement of Ig genes in non B-cells”).
V(D)J Rearrangement Patterns
In the mouse and human systems the rearrangement of Ig H and L chains is often described as an ordered, regulated program.49 The well-studied steps involve formation of the DJ before recombination of VH to the DJ in pro B-cells, followed by cell division and subsequent rearrangement of the L chain genes in pre B-cells, where the kappa L chain (Igκ) locus is activated before the second L chain isotype, lambda (Igλ). The regulated accessibility of different genes and gene segments to RAG enable one H chain allele to be expressed (allelic exclusion, H chain exclusion) with one allele of either κ or λ L chain (allelic and isotypic exclusion); hence, one kind of antigen receptor per lymphocyte. Outside of the mouse model there is currently little information on TCR or Ig chromatin and DNA modification, but V(D)J rearrangement patterns reflect the order of gene accessibility to RAG and these are compared among mouse, rabbit, chicken and shark.
Mouse
In the mouse IgH gene rearrangement takes place in a set order and in step-wise fashion.59,51 At the pro B-cell stage the chromatin domain encompassing the D, JH and Cμ genes become activated, probably through the intronic enhancer and allow D to JH recombination on both chromosomes. This is followed by activation of the chromatin domain containing the upstream VH genes. Because of the very large distance between the VH gene segments and the DJ, locus contraction and looping of the DNA52-54 are required to bring them into close proximity for rearrangement (Fig. 2). H chain exclusion is the outcome of the staggering of the V to DJ step between the two alleles. If the first VDJ is not viable, rearrangement continues on the homologous chromosome.
The initiation and maintenance of allelic exclusion involves relocation of the genes in nuclear compartments.53,55 In pro B-cells IgH repositions away from the nuclear periphery and this may have to do with its activation; in pre B-cells the nonrearranged allele is recruited to the pericentrometric heterochromatin, an interaction thought to be repressive for recombination. How rearrangement begins at one allele before the other is not clear and the basis may differ at the IgH, TCRβ and Igκ genes. An explanation for asynchronous rearrangement at the TCRβ locus has been recently proposed after finding that in rearranging T-cells both alleles of TCRβ interacted with repressive nuclear compartments at equal and high frequency.56 This observation suggests a limited window of opportunity to achieve the V to DJ step and that any rearrangement is consequently a very low frequency event. Two simultaneous rearrangements in a cell are thus unlikely to occur and allelic inclusion is avoided.
Rabbit
Mouse IgH configurations in hybridoma cell lines57 reflect the frequency of the recombination events, which render 51% of them VDJ/DJ, 44% VDJ/VDJ- and 5% VDJ/germline (VDJ is the expressed rearrangement, VDJ- is nonfunctional). In contrast, the IgH configurations in rabbit cell lines were: 40% VDJ/DJ, 10% VDJ/VDJ- and 50% VDJ/germline.58 Since the D to JH step occurred on both alleles in 95% of mouse B-cells, the finding that it has not done so in 50% of rabbit B-cells suggests that asynchrony between IgH alleles can exist to a greater extent in rabbit. Lanning and coworkers59 hypothesized that the D to JH rearrangement in rabbit involves slower kinetics and is the rate-determining step; once DJ is achieved on one chromosome there is rapid recombination to VDJ. Because of the overall inefficiency of the D to JH step, the relatively few numbers of cells with VDJ/VDJ- reflect a restricted time opportunity for the laggard allele to achieve VDJ.
Although some infrequent VH to D rearrangement was observed in rabbit splenocytes,58 its significance is unknown, since the recombined DJ is the primary intermediate isolated from pro-B-cells. Cloned fetal rearrangements carried the two D-proximal VH genes, VH1 and the neighboring pseudogene VH2,60 despite > 100 available VH upstream; this early rearrangement bias together with clonal expansion of B-cells with VH1-expressing VDJ causes such H chains to be 70-90% of expressed Ig molecules.61 Usage of the D-proximal VH1 in rabbit can be likened to the preferential rearrangement of the D-proximal VH genes in fetal mouse liver,62,63 but the molecular basis of either remains be to elucidated.
Chicken
The earliest recombined cells are in the yolk sac at day 5 and 6 of incubation and carry DJ only; VDJ is found on day 9.64 Rearrangement occurs exclusively to the D-proximal VH gene, the only functional gene out of multiple VH elements; the other VH act as donor templates during the gene conversion process in the bursa. The primary Ig repertoire in chicken, as in rabbits, is generated by postrearrangement gene conversion.65,62 There is a distinct DJ/DJ step that is B-lineage specific in chicken and this is followed by simultaneous V rearrangement at the H and L chain loci.64,66 Clones carrying only the VJ or only the VDJ could be observed,66 showing that there is no ordered H and L chain rearrangement, as there exists in mouse and rabbit.61 Thus, in the chicken, L chain rearrangement is not dependent on the success of H chain rearrangement and there is no pre-B-cell stage as in mammals.
More than 90% of bursal follicles contain the VDJ/DJ configuration and none carried VDJ rearrangements on both chromosomes. Similarly, only one allele of the L chain recombined. It was suggested that the V rearrangement occurs after removal of repression from one allele randomly and that this is an event of such low efficiency that there is little probability of its occurrence on both alleles.67
Multiple IgH Loci in Other Vertebrate Species
The contrasting examples of mouse, rabbit and chicken show that the V(D)J recombination program is adapted for each species. There is at least one step that is limited by RAG accessibility and/or time constraints67 and the factors that determine these parameters remain to be elucidated. These three systems all involve a choice of two H chain alleles, but when one recombination step tends to be limiting or occurring at very low frequency, then the presence of additional alleles—one or two, equally subjected to the constraints—would not greatly increase the chances for allelic inclusion. Model systems genetically manipulated to carry multiple H chain genes (interspecies hybrid tetraploid and triploid Xenopus68 and mice triallelic for IgH69) do exhibit monoallelic H chain expression and thus the same would be expected for those animals with more than one naturally-occurring IgH locus. Polyploid Xenopus species carry multiple active IgH genes.70 Bony fish, alone of all vertebrate classes, underwent an additional genome-wide duplication71 and some species support more than one IgH locus although in most only one remains.
Ig Rearrangement in the Shark
The IgH minilocus organization in cartilaginous fishes, representatives of the earliest vertebrates, is considered primitive and ancestral to the classical IgH locus in other vertebrates. Sharks, rays and skates carry 15-200 miniloci (“clusters”) each consisting of a few gene segments (VH-D1-D2-JH-Cμ)4,23 as shown in (Fig. 2). In most species the rearranging elements are located within a total span of 2 kb. The clusters themselves are located far apart from each other,72 >120 kb and can be situated on different chromosomes.73 V(D)J recombination takes place among the four gene segments of the minilocus; there is no evidence for intercluster rearrangement in B-cells and hence no need for locus contraction in such a system. The close proximity of the gene segments (400 bp apart) also makes unlikely any separately activated chromatin domains within a cluster. In fact, there is no strict order of rearrangement of the VH, D1, D2 and JH. Once an IgH gene is activated in a precursor B-cell, its gene segments recombine all at once and to completion.74
In single B-cell studies, few Ig transcripts75 and few genomic rearrangements74 were observed per lymphocyte. In the nurse shark there are 9-12 functional IgH genes and in any B-cell there are 1-3 VDJ genomic rearrangements of which only one appeared to encode a viable receptor. Less than 10% of the cells carried any partially rearranged genes and the rest of the IgH genes were in germline configuration. This suggests that once initiated, recombination occurs efficiently between the four gene segments. These data show that H chain exclusion exists in the shark, despite its unique IgH organization. As in higher vertebrates, H chain exclusion in sharks is based on limitation of rearrangement, but the mechanism of repression (or activation) must accommodate the large and varied numbers of IgH loci in different cartilaginous fish species.
The process producing monoallelic Ig H chain expression at the murine IgH locus evolved with and is a consequence of the complex gene organization, whose multiple gene segments are scattered over 2 Mb. If you take away the locus contraction and the separately activated domains, what shark and mouse have in common is that initiation of rearrangement is an inefficient, low frequency event. Whether there are regulatory features in common between shark and tetrapod IgH gene systems remains to be established. However, a few conclusions can be extracted. Because of the large number of IgH loci and their dispersed locations, it is unlikely that H chain exclusion in the shark is based on any mechanism that predetermines77,78 rearrangement preference at homologous chromosomes. In nurse shark at least two IgH genes are adjacent72 and the model for kappa L chain exclusion based on rearrangement preference evinced by the earlier replicating chromosome will not distinguish multiple, linked genes. It is not clear whether the 1-3 rearrangements in a B-cell occurred simultaneously or sequentially and we suggest that their activation was probably stochastic. If it happened that one rearrangement at a shark IgH gene is nonfunctional it seems unlikely that its allele is more apt to be the one next (or simultaneously) targeted for recombination than an adjacent or any other IgH in the genome.
Rearrangement of Ig Genes in Non B-cells
There exist pre-rearranged Ig genes in the germline of cartilaginous fishes, catfish and chicken.78-80 In sharks, skates and rays some IgH clusters carry partially or fully recombined VD-J or VDJ and the IgL clusters joined VJ.4 Examination of nurse shark L chain junctions in some germline-joined VJ showed P region sequence that may indicate a one-time hairpin formation. This evidence and the fact that the “12/23 rule” is always obeyed, suggest that there was RAG activity in germ cells of some animals.81,19 It was hypothesized that RAG-mediated changes in germline Ig genes produced the VD templates used in chicken H chain gene conversion or perhaps generated D elements during antigen receptor gene evolution.
The function of recombined genes in the shark antibody repertoire is not known; it appears that many are pseudogenes. In a species with many pre-rearranged VDJ there would be a strong likelihood for allelic inclusion if more than one IgH is activated at a time, but at the moment the germline genes in these animals have not been fully characterized. Nurse shark is an instance where all its IgM clusters have been characterized and none are pre-rearranged, showing that germline-joined genes are particular to the species.72
Once initiated, somatic rearrangement in B-cells leads to VDJ. Partially rearranged IgH on the other hand have been observed in abundance in nurse shark thymocytes and 3-7 can be isolated per cell.74 Thymic H chain transcripts could not be detected, implying that availability of DNA to RAG does not require transcription. That many thymocyte rearrangements are incomplete as VD-D-J, V-DDJ, etc., suggests that transcription may be part of the process that recruits82 RAG to its target for efficient recombination. This IgH rearrangement-permissive state in thymocytes may have characteristics in common with that in germ cells enabling RAG, when present, to effect recombination. However, the state of the IgH chromatin in either cell type has yet to be characterized.
About L Chain
In the course of evolution, whole-locus duplications produced the multiple cluster organization of cartilaginous fish IgH and IgL, whereas successive tandem duplications of the gene segments V, (D) and J generated the “translocon” organization that exists in tetrapods. While H chain genes are organized either as translocon or multiple clusters, the evolution of L chain genes83 is more complex.
The number of L chain isotypes varies among vertebrates. In chicken there is only the one locus, Igλ; in mammals there are two, Igλ and Igκ. In Xenopus there are three: Igσ (sigma) and the homologs of Igκ (Igρ, called rho) and Igλ (called type III). In shark there are four: cartilaginous fish-specific “Igσ-cart” (called type I/NS5) and the homologs of sigma, Igκ (called type III/NS4) and Igλ (called type II/NS3). Igκ is thus present in all animals except birds and its organization varies considerably. In tetrapods Igκ is one locus. In nurse shark the Igκ homolog exists as >60 miniloci, with one V, one J and one C exon and tend to be separated by some distance. However in a bony fish like zebrafish, Igκ genes (type 1/3)84 are arranged closely in serial arrays (examples in Fig. 4) and on at least four different chromosomes.85
It is not clear how L chain expression is regulated in zebrafish (or any bony fish). In cod it was shown that multiple enhancers existed in the serial clusters but not every IgL C region was associated with downstream enhancer activity.86 It cannot be anticipated from mere distance how regulatory control is exercised. Because there can be additional possibilities for intracluster rearrangement following an initial V to J attempt (Fig. 4), we have suggested that the bony fish organization allows for correction not only of nonproductive VJ but also in-frame VJ that contribute to forming a self-reactive specificity.84 In other words, there exists a potential for receptor editing87 in fishes, since the organizational set-up appears to allow for secondary rearrangements.
In zebrafish, the IgH organization is translocon like tetrapods88 so that both types of arrangement exist for its Ig genes. It is clear that H and L chain gene organizations do not have to co-evolve—as they did not in bony fish89—and information from this and the other model systems suggest they can be regulated independently. L chain exclusion is not as stringent as H chain, but the mechanism for restricting their expression in zebrafish must manage a large array of clusters, many of which carry multiple recombinogenic elements on either side of the C exon. How V(D)J recombination is sorted out in zebrafish will elucidate those aspects of RAG accessibility that evolve with individual species' immune system requirements.
Summary
V(D)J rearrangement was established in an ancestral jawed vertebrate about 500 million years ago. From sharks to mammals two features are evolutionarily conserved—the mechanism of RAG recombinase action and a process for limiting rearrangement activity in order to produce monospecific lymphocytes. The regulated accessibility of antigen receptor genes to RAG was characterized in precursor lymphocytes of mice and human beings, where it is usually described as an ordered, two-stage program. However, a comparison of Ig rearrangement patterns from rabbit, chicken and shark shows that this process neither has to be strictly ordered nor must take place in two stages to generate a diverse repertoire and bring about allelic exclusion.
Acknowledgments
I wish to thank Louis Du Pasquier and Martin Flajnik for reading the manuscript and Karolina Malecek for her help with the figures. This work was supported by grants from the National Institutes of Health and the National Science Foundation.
References
- 1.
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. [PubMed: 6300689]
- 2.
- Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. [PubMed: 2598259]
- 3.
- Oettinger MA, Schatz DG, Gorka C. et al. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. [PubMed: 2360047]
- 4.
- Rast JP, Litman GW. Towards understanding the evolutionary origins and early diversification of rearranging antigen receptors. Immunol Rev. 1998;166:79–86. [PubMed: 9914904]
- 5.
- Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol. 2004;25:640–644. [PubMed: 15530832]
- 6.
- Spring J. Genome duplication strikes back. Nature Genetics. 2002;31:128–129. [PubMed: 12040374]
- 7.
- Furlong RF, Holland PWH. Were vertebrates octoploid? Philos Trans R Soc Lond B Biol Sci. 2002;357:531–544. [PMC free article: PMC1692965] [PubMed: 12028790]
- 8.
- Holland PWH, Garcia-Fernandez J, Williams NA. et al. Gene duplications and the origins of vertebrate development. Development Suppl. 1994:125–133. [PubMed: 7579513]
- 9.
- Escriva H, Manzon L, Youson J. et al. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol biol Evol. 2002;19:1440–14506. [PubMed: 12200472]
- 10.
- Kasahara M, Suzuki T, Du Pasquier L. On the origins of the adaptive immune system: novel insights from invertebrates and cold-blooded vertebrates. Trends Immunol. 2004;25:105–111. [PubMed: 15102370]
- 11.
- Sakano H, Huppi K, Heinrich G. et al. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. [PubMed: 111144]
- 12.
- van Gent DC, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. [PubMed: 8599117]
- 13.
- Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. [PMC free article: PMC1131882] [PubMed: 15898832]
- 14.
- Fugmann SD, Messier C, Novack LA. et al. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci USA. 2006;103:3728–3733. [PMC free article: PMC1450146] [PubMed: 16505374]
- 15.
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. [PubMed: 9723614]
- 16.
- Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. [PubMed: 9727489]
- 17.
- Thompson CB. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. [PubMed: 7584143]
- 18.
- Fugmann SD, Lee AI, Shockett PE. et al. The RAG proteins and V(D)J recombination: complexes, ends and transposition. Annu Rev Immunol. 2000;18:495–527. [PubMed: 10837067]
- 19.
- Lewis SM, Wu GE. The old and the restless. J Exp Med. 2000;191:1631–1636. [PMC free article: PMC2193153] [PubMed: 10811857]
- 20.
- Greenberg AS, Avila D, Hughes M. et al. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. [PubMed: 7877689]
- 21.
- Criscitiello MF, Saltis M, Flajnik MF. An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci USA. 2006;103:5036–5041. [PMC free article: PMC1458790] [PubMed: 16549799]
- 22.
- Parra ZE, Baker ML, Schwarz RS. et al. A unique T-cell receptor discovered in marsupials. Proc Natl Acad Sci USA. 2007;104:9776–9781. [PMC free article: PMC1887558] [PubMed: 17535902]
- 23.
- Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol. 2002;2:688–698. [PubMed: 12209137]
- 24.
- Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Annu Rev Immunol. 1999;17:109–147. [PubMed: 10358755]
- 25.
- Rast JP, Anderson MK, Strong SJ. et al. α, β, γ and δ T-cell antigen receptor genes arose early in vertebrate phylogeny. Immunity. 1997;6:1–11. [PubMed: 9052832]
- 26.
- Wilson MR, Zhou H, Bengtén E. et al. T-cell receptors in channel catfish: structure and expression of TCR alpha and beta genes. Mol Immunol. 1998;35:545–557. [PubMed: 9809582]
- 27.
- Haire RN, Kitzan Haindfield MK. et al. Structure and diversity of T-lymphocyte antigen receptors alpha and gamma in Xenopus. Immunogenetics. 2002;54:431–438. [PubMed: 12242593]
- 28.
- André S, Kerfourn F, Affaticati P. et al. Highly restricted diversity of TCR delta chains of the amphibian Mexican axolotl (Ambystoma mexicanum) in peripheral tissues. Eur J Immunol. 2007;37:1621–1633. [PubMed: 17523213]
- 29.
- Kubota T, Wang J, Göbel TW. et al. Characterization of an avian (Gallus gallus domesticus) TCR alpha delta gene locus. J Immunol. 1999;163:3858–3866. [PubMed: 10490985]
- 30.
- Parra ZE, Baker ML, Hathaway J. et al. Comparative genomic analysis and evolution of the T-cell receptor loci in the opossum Monodelphis domestica. BMC Genomics. 2008;9:111. [PMC free article: PMC2275272] [PubMed: 18312668]
- 31.
- Parra ZE, Arnold T, Nowak MA. et al. TCR gamma chain diversity in the spleen of the duckbill platypus (Ornithorhynchus anatinus). Dev Comp Immunol. 2006;30:699–710. [PubMed: 16303181]
- 32.
- Krangel MS. Gene segment selection in V(D)J recombination: accessibility and beyond. Nat Immunol. 2003;4:624–630. [PubMed: 12830137]
- 33.
- Gellert M. V(D)J recombination: RAG proteins, repair factors and regulation. Annu Rev Biochemistry. 2002;71:101–132. [PubMed: 12045092]
- 34.
- Komori T, Okada A, Stewart V. et al. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. [PubMed: 8356451]
- 35.
- Gilfillan S, Dierich A, Lemeur M. et al. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. [PubMed: 8356452]
- 36.
- Bertocci B, De Smet A, Weill JC. et al. Nonoverlapping functions of DNA polymerases mu, lambda and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. [PubMed: 16860755]
- 37.
- Domínguez O, Ruiz JF, Laín de Lera T. et al. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. [PMC free article: PMC310241] [PubMed: 10747040]
- 38.
- Beetz S, Diekhoff D, Steiner LA. Characterization of terminal deoxynucleotidyl transferase and polymerase mu in zebrafish. Immunogenetics. 2007;59:735–744. [PubMed: 17701034]
- 39.
- Zhang SM, Adema CM, Kepler TB. et al. Diversification of Ig superfamily genes in an invertebrate. Science. 2004;305:251–254. [PubMed: 15247481]
- 40.
- Wu TT, Johnson G, Kabat EA. Length distribution of CDRH3 in antibodies. Proteins. 1993;16:1–7. [PubMed: 8497480]
- 41.
- Fleurant M, Changchien L, Chen CT. et al. Shark Ig light chain junctions are as diverse as in heavy chains. J Immunol. 2004;173:5574–5582. [PubMed: 15494507]
- 42.
- Wedemayer GJ, Patten PA, Wang LH. et al. Structural insights into the evolution of an antibody combining site. Science. 1997;276:1665–1669. [PubMed: 9180069]
- 43.
- Wilson IA, Stanfield RL. Antibody-antigen interactions: new structures and new conformational changes. Curr Opin Struct Biol. 1994;4:857–867. [PubMed: 7536111]
- 44.
- Stanfield RL, Dooley H, Verdino P. et al. Maturation of shark single-domain (IgNAR) antibodies: evidence for induced-fit binding. J Mol Biol. 2007;367:358–372. [PubMed: 17258766]
- 45.
- Nguyen VK, Hamers R, Wyns L. et al. Camel heavy-chain antibodies: diverse germline V(H)H and specific mechanisms enlarge the antigen-binding repertoire. EMBO J. 2000;19:921–930. [PMC free article: PMC305632] [PubMed: 10698934]
- 46.
- Achour I, Cavelier P, Tichit M. et al. Tetrameric and homodimeric camelid IgGs originate from the same IgH locus. J Immunol. 2008;181:2001–2009. [PubMed: 18641337]
- 47.
- Burnet FM. The clonal selection theory of acquired immunity. Cambridge: Cambridge University Press. 1959
- 48.
- Stanhope-Baker P, Hudson KM, Shaffer AL. et al. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–897. [PubMed: 8681383]
- 49.
- Yancopoulos GD, Alt FW. Regulation of the assembly and expression of variable-region genes. Annu Rev Immunol. 1986;4:339–368. [PubMed: 3085692]
- 50.
- Alt FW, Yancopoulos GD, Blackwell TK. et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. [PMC free article: PMC557501] [PubMed: 6086308]
- 51.
- Chowdhury D, Sen R. Regulation of immunoglobulin heavy-chain gene rearrangements. Immunol Rev. 2004;200:182–196. [PubMed: 15242405]
- 52.
- Fuxa M, Skok J, Souabni A. et al. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. [PMC free article: PMC359395] [PubMed: 15004008]
- 53.
- Kosak ST, Skok JA, Medina KL. et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. [PubMed: 11935030]
- 54.
- Jhunjhunwala S, van Zelm MC, Peak MM. et al. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. [PMC free article: PMC2771211] [PubMed: 18423198]
- 55.
- Roldán E, Fuxa M, Chong W. et al. Locus “decontraction” and centromic recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat Immunol. 2005;6:31–41. [PMC free article: PMC1592471] [PubMed: 15580273]
- 56.
- Schlimgen RJ, Reddy KL, Singh H. et al. Initiation of allelic exclusion by stochastic interaction of Tcrb alleles with repressive nuclear compartments. Nat Immunol. 2008;9:802–809. [PMC free article: PMC2561338] [PubMed: 18536719]
- 57.
- Perlot T, Alt FW, Bassing CH. et al. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci USA. 2005;102:14362–4367. [PMC free article: PMC1242331] [PubMed: 16186486]
- 58.
- Tunyaplin C, Knight KL. IgH gene rearrangements on the unexpressed allele in rabbit B-cells. J Immunol. 1997;158:4805–4811. [PubMed: 9144495]
- 59.
- Lanning D, Jasper P, Knight K. IgH haplotype exclusion in rabbits. Semin Immunol. 2002;14:163–168. [PubMed: 12160644]
- 60.
- Tunyaplin C, Knight KL. Fetal VDJ gene repertoire in rabbit: evidence for preferential rearrangement of VH1. Eur J Immunol. 1995;25:2583–2587. [PubMed: 7589130]
- 61.
- Mage RG, Lanning D, Knight KL. B-cell and antibody repertoire development in rabbits: the requirement of gut-associated lymphoid tissues. Dev Comp Immunol. 2006;30:137–153. [PubMed: 16098588]
- 62.
- Yancopoulos GD, Desiderio SV, Paskind M. et al. Preferential utilization of the most JH-proximal VH gene segments in preB-cell lines. Nature. 1984;311:727–733. [PubMed: 6092962]
- 63.
- Knight KL, Becker RS. Molecular basis of the allelic inheritance of rabbit immunoglobulin VH allotypes: implications for the generation of antibody diversity. Cell. 1990;60:963–970. [PubMed: 2317867]
- 64.
- Reynaud CA, Imhof BA, Anquez V. et al. Emergence of committed B-lymphoid progenitors in the developing chicken embryo. EMBO J. 1992;11:4349–4358. [PMC free article: PMC557008] [PubMed: 1425572]
- 65.
- Weill JC, Reynaud CA. The chicken B-cell compartment. Science. 1987;238:1094–1098. [PubMed: 3317827]
- 66.
- Benatar T, Tkalec L, Ratcliffe MJ. Stochastic rearrangement of immunoglobulin variable-region genes in chicken B-cell development. Proc Natl Acad Sci USA. 1992;89:7615–7619. [PMC free article: PMC49761] [PubMed: 1502173]
- 67.
- Weill JC, Cocea L, Reynaud CA. Allelic exclusion: lesson from GALT species. Semin Immunol. 2002;14:213–215. [PubMed: 12160649]
- 68.
- Du Pasquier L, Hsu E. Immunoglobulin expression in diploid and polyploid interspecies hybrid of Xenopus: evidence for allelic exclusion. Eur J Immunol. 1983;13:585–590. [PubMed: 6873155]
- 69.
- Barreto V, Meo T, Cumano A. Mice triallelic for the Ig heavy chain locus: implications for VHDJH recombination. J Immunol. 2001;166:5638–5645. [PubMed: 11313404]
- 70.
- Du Pasquier L, Blomberg B. The expression of antibody diversity in natural and laboratory-made polyploid individuals of the clawed toad Xenopus. Immunogenetics. 1982;15:251–60. [PubMed: 6802750]
- 71.
- Hoegg S, Brinkmann H, Taylor JS. et al. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59:190–203. [PubMed: 15486693]
- 72.
- Lee V, Huang JL, Lui MF. et al. The evolution of multiple isotypic IgM heavy chain genes in the shark. J Immunol. 2008;180:7461–7470. [PMC free article: PMC2590587] [PubMed: 18490746]
- 73.
- Anderson M, Amemiya C, Luer C. et al. Complete genomic sequence and patterns of transcription of a member of an unusual family of closely related, chromosomally dispersed Ig gene clusters in Raja. Int Immunol. 1994;6:1661–1670. [PubMed: 7865459]
- 74.
- Malecek K, Lee V, Feng W. et al. Immunoglobulin heavy chain exclusion in the shark. PLoS Biol. 2008;6:e157. [PMC free article: PMC2435157] [PubMed: 18578572]
- 75.
- Eason DD, Litman RT, Luer CA. et al. Expression of individual immunoglobulin genes occurs in an unusual system consisting of multiple independent loci. Eur J Immunol. 2004;34:2551–2558. [PubMed: 15307187]
- 76.
- Mostoslavsky R, Singh N, Kirillov A. et al. Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12:1801–1811. [PMC free article: PMC316908] [PubMed: 9637682]
- 77.
- Mostoslavsky R, Singh N, Tenzen T. et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. [PubMed: 11700561]
- 78.
- Kokubo F, Litman R, Shamblott MJ. et al. Diverse organization of immunoglobulin VH gene loci in a primitive vertebrate. EMBO J. 1988;7:3413–3422. [PMC free article: PMC454840] [PubMed: 3145194]
- 79.
- Ghaffari SH, Lobb CJ. Structure and genomic organization of a second cluster of immunoglobulin heavy chain gene segments in the channel catfish. J Immunol. 1999;162:1519–1529. [PubMed: 9973409]
- 80.
- Reynaud CA, Dahan A, Anquez V. et al. Somatic hyperconversion diversifies the single VH gene of the chicken with a high incidence on the D region. Cell. 1989;40:283–291. [PubMed: 2507167]
- 81.
- Lee SS, Fitch D, Flajnik MF. et al. Rearrangement of immunoglobulin genes in shark germ cells. J Exp Med. 2000;191:1637–1648. [PMC free article: PMC2193156] [PubMed: 10811858]
- 82.
- de Villartay J-P. Passera ou ne passera pas—accessibility is key. Nature Immunol. 2006;7:1019–1021. [PubMed: 16985496]
- 83.
- Criscitiello MF, Flajnik MF. Four primordial immunoglobulin light chain isotypes, including lambda and kappa, identified in the most primitive living jawed vertebrates. Eur J Immunol. 2007;37:2683–2694. [PMC free article: PMC7094790] [PubMed: 17899545]
- 84.
- Hsu E, Criscitiello MF. Diverse immunoglobulin light chain organizations in fish retain potential to revise B-cell receptor specificities. J Immunol. 2006;177:2452–2462. [PMC free article: PMC3129705] [PubMed: 16888007]
- 85.
- Zimmerman AM, Yeo G, Howe K. et al. Immunoglobulin light chain (IgL) genes in zebrafish: Genomic configurations and inversional rearrangements between (VL-JL-CL) gene clusters. Dev Comp Immunol. 2008;32:421–434. [PMC free article: PMC3014032] [PubMed: 18022691]
- 86.
- Bengtén E, Stromberg S, Daggfeldt A. et al. Transcriptional enhancers of immunoglobulin light chain genes in Atlantic cod (Gadus morhua). Immunogenetics. 2000;51:647–658. [PubMed: 10941836]
- 87.
- Nemazee D. Receptor editing in lymphocyte development and central tolerance. Nat Rev Immunol. 2006;6:728–740. [PubMed: 16998507]
- 88.
- Danilova N, Bussmann J, Jekosch K. et al. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol. 2005;6:295–302. [PubMed: 15685175]
- 89.
- Daggfeldt A, Bengtén E, Pilstrom L. A cluster type organization of the loci of the immunoglobulin light chain in Atlantic cod (Gadus morhua L.) and rainbow trout (Oncorhynchus mykiss Walbaum) indicated by nucleotide sequences of cDNAs and hybridization analysis. Immunogenetics. 1993;38:199–209. [PubMed: 8505063]
Figures
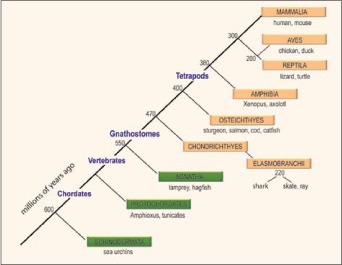
Figure 1
Evolution of the chordates. The phylogenetic relationships among chordates is shown (boxes) with notations of the major animal models in each taxon beneath the boxes. The adaptive immune system, defined by RAG-mediated rearranging antigen receptor genes of the Ig superfamily and by the major histocompatibility complex, has been found only in the jawed vertebrates (gnathostomes, beige boxes). Protochordates include Cephalochordates (Amphioxus) and Urochordates (Ciona/tunicates). Numbers denote when the taxa emerged in evolution (millions of years ago). Reprinted with some alterations; Hsu E, Pulham N, Rumfelt LL et al. The plasticity of immunoglobulin gene systems in evolution. Immunol Rev 2006; 210:8-26. Copyright Blackwell Munksgaard 2006.
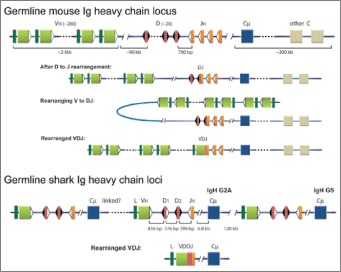
Figure 2
Comparison of Ig H chain genes in mouse and shark. Germline mouse Ig H chain locus: the mammalian H chain locus consists of a series of tandemly duplicated VH, D and JH gene segments that rearrange during B-cell development. The recombined VDJ is transcribed with one of the downstream constant (C) region genes, here simplified as single units (blue box is Cμ ). The VH is represented by olive boxes, preceded by the leader sequence in dark green and flanked by the recombination signal sequence (RSS, white triangle) at the 3′ end, that consists of heptamer and nonamer motifs separated by a 23 bp spacer sequence. As indicated, the distance between the 3′-most VH and the first functional D is 90 kb. The D gene segments in red, flanked on both sides by RSS (black triangles) containing 12 bp spacers and the JH gene segments (orange) with 23 bp spacer RSS. After D to J rearrangement: the first stage of rearrangement involves recombination between D and JH, with the intervening DNA excised. The DJ product is depicted as a fusion of the red and orange boxes, with the RSS flanking its 5′ end. Rearranging V to DJ: locus contraction and looping of the DNA allows linearly distant VH gene segments to recombine with the DJ. The final VDJ product is shown as Rearranged VDJ. Germline shark Ig H chain loci: the IgM H chain genes in sharks and skates (cartilaginous fishes) are multiple miniloci each consisting of VH, two D, one JH and one Cμ gene (blue box). The gene segments in any nurse shark IgH gene are located about 400 bp apart as shown but are distant (e.g., 6.3-6.8 kb) from the Cμ1 exon. The physical relationships among the loci are not clear except for one instance, where they were located 120 kb apart.72 Rearranged VDJ: the four gene segments rearrange within the minilocus to VDDJ (called VDJ). Whereas in mouse IgH gene rearrangement takes place in a strict order (D to JH before VH to DJ), the rearrangement of the four gene segments in the shark takes place at once and without any strict order. Reprinted with permission from Malecek K, Lee V, Feng W et al. Immunoglobulin heavy chain exclusion in the shark. PLoS Biol 2008; 6:e157. Copyright 2008 Malecek et al. A color version of this image is available at www.eurekah.com
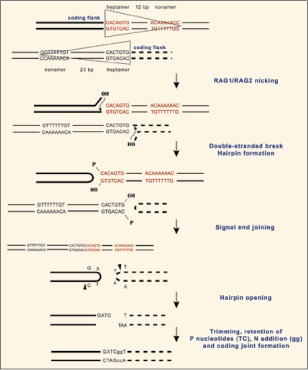
Figure 3
RAG-mediated recombination. Details of V(D)J recombination are described in other chapters of this book. The flanks of gene segments before rearrangement are shown with the RSS enclosed by triangles, to correlate with the symbols in Figures 2 and 4. The RSS pair is bound by RAG, which introduces nicks. The nicking occurs 5′ of the 7-mer end of the RSS on the top strand of each of the two Ig gene segments, producing a 3′-hydroxy on the coding end of the Ig gene segment and a 5′ phosphoryl on the RSS (signal) end. The result is a duplex nicked at either RSS. The second step involves intramolecular transesterfication reactions where the 3′-OH attack the opposing phosphodiester bonds, causing the coding ends to become a covalently closed hairpins and freeing the blunt signal ends. Joining of the ends is carried out by the nonhomologous end joining repair pathway. The hairpin coding ends are opened asymmetrically by the nuclease Artemis and the resultant single-stranded overhang consists of a portion of the coding end and its complementary sequence. Sometimes the overhang could be included as part of the final joined product and is observed as inverted repeat sequence (P region). The DNA ends are trimmed; TdT may insert nontemplated nucleotides (lower case letters). Reprinted with some alterations; Hsu E. Immunoglobulin recombination signal sequences: somatic and evolutionary functions. In: Caporale L, ed. The Implicit Genome, New York: Oxford University Press, 2005, Chapter 9. Copyright 2006 by Oxford University Press, Inc.
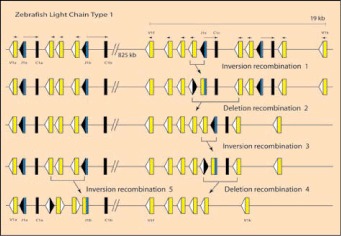
Figure 4
Organization of representative genes encoding zebrafish L chains. Some L chain type 1 clusters on chromosome 24 are represented on top line; the names of segments are some of those identified in reference 84; their updated linkage, polarity and distances were obtained from the Zv7 zebrafish genome assembly (www.ensembl.org) and reference 85. V (yellow boxes) and J (blue) gene segments are flanked by RSS (white triangle is RSS with 12 bp spacer, black triangle is RSS with 23 bp spacer) and C exons are depicted by black boxes. The transcriptional polarities are indicated by overhead arrows. A hypothetical series of rearrangements is depicted. Inversion recombination 1: rearrangement between J1c and V1i immediately upstream to form VJ (indicated as fused rectangles) and blunt-end joined RSS (fused triangles). Deletion recombination 2: The RSS-23 of the fused signal joint recombines with downstream V gene segment and deletes intervening DNA. Inversion recombination 3: the remaining J rearranges to upstream V, forming again VJ and blunt-end joined RSS. This VJ can be excised by deletion recombination 4 and replaced by rearrangement at another cluster, inversion recombination 5. A color version of this image is available at www.eurekah.com