NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Secretion of proteins from eukaryotic cells requires the coordinated function of multiple organelles and cellular machineries. After synthesis and translocation into the endoplasmic reticulum, proteins are exported to the Golgi apparatus, a multi-compartment organelle that is the protein modifying, packaging and distribution center of the secretory pathway. This chapter provides a brief historical account of the discovery of the Golgi apparatus, a description of its unique structure and organization, and its role in glycoprotein biosynthesis, sorting and secretion. The biogenesis of the Golgi through localization of resident proteins and its inheritance during cell division is also described. Emphasis is placed on processes, such as protein transport through the Golgi, that are incompletely understood and remain focal points for current research in this field.
Introduction
The Secretory Pathway
The transport of proteins and lipids from their site of synthesis at the endoplasmic reticulum (ER) to the cell surface is mediated by the secretory pathway and is an essential process in eukaryotic organisms. A great variety of molecules are extruded from cells by the action of the secretory pathway, including extracellular matrix components that provide the foundation for constructing tissues and organs. Moreover, this pathway plays a major role in the biogenesis of the plasma membrane and its expansion before cell division. Therefore, without secretion there would be no cells, tissues or organs, and so it is safe to say that we owe our very existence to the secretory pathway.
To understand the process of secretion we must learn about the organelles that compose the secretory pathway; the ER and Golgi apparatus, and the transport vesicles these organelles produce. The membrane of these organelles is primarily synthesized and assembled at the ER but with contributions from mitochondria (phosphatidylethanolamine) and the Golgi apparatus (sphingolipids). Newly synthesized proteins destined for secretion gain entry into the secretory pathway by translocation across the ER membrane. This translocation apparatus also integrates proteins into the membrane and establishes their topology with respect to the lipid bilayer (see Chapter 7). Many secretory proteins are covalently modified with oligosaccharides to produce glycoproteins, a biosynthetic process initiated in the ER and continued in the Golgi apparatus. Once proteins are properly folded and modified in the ER, they are allowed to leave and are ushered into COPII-coated carrier vesicles forming at specific exit sites (see Chapters 1 and 8). After budding, the COPII vesicles uncoat and deliver their contents to an amazing protein modifying, transporting and sorting machine called the Golgi apparatus, which will be the focus of this chapter. From the Golgi, proteins can be delivered back to the ER, to the endosomal-lysosomal system or to the plasma membrane in different membrane-bound carriers.
History of the Golgi Apparatus
Nature must be amused at our attempts to understand the Golgi apparatus. This organelle has been steeped in controversy for more than 100 years and will likely continue this fine tradition for years to come. It was discovered by its namesake, Camillo Golgi, and was first described in 1898 as an intracellular reticular apparatus stained by the “black reaction” in neuronal cells.1 This histochemical stain is caused by the reduction of silver nitrate or osmium in compartments of the Golgi apparatus. In the early 1900s, Golgi, Negri, Cajal and others used this method to visualize a similar reticular apparatus in many different cells,1-3 leading to the current view that the Golgi apparatus is ubiquitous in eukaryotic cells. This intracellular entity was initially known as the “reticular structure of Golgi”, although the term “Golgi apparatus”, first used by Nusbaum in 1913, has slowly become the most popular name for this organelle.4 Fuchs first speculated on a role for the Golgi apparatus in protein secretion in 1902,5 and the morphological studies of Nassonov6 and Bowen7 strongly supported this opinion. However, many detractors, such as Baker, argued that the Golgi apparatus was simply an artifact of the staining method used to observe it, and was perhaps no more than the deposition of metal in “empty spaces” between other cellular structures.8 This debate raged until the 1950s when the Golgi apparatus was visualized by Felix and Dalton using the newly developed electron microscope.9,10 While others pointed out similar structures in papers published concomitantly, 11,12 it was Felix and Dalton that demonstrated the osmiophilic nature of the organelle, correlating it to the Golgi apparatus characterized by light microscopy after impregnation with OsO4. These authors also introduced the term “Golgi complex” to emphasize its multi-component nature and this name is still favored by many cell biologists.10 A more complete accounting of the Golgi apparatus history can be found in a few excellent books and reviews (refs. 4,13-16) .
Structure of the Golgi Apparatus
Cisternal Organization
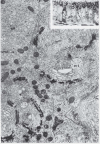
As visualized by electron microscopy (EM) of ultrathin sections, the Golgi apparatus appears as a stack containing multiple flattened, disk-shaped membranes called cisternae (Fig. 1). To interpret these images, it helps to imagine a stack of pancakes (cisternae), cut down the center, and viewed from the edge of the cut face of the stack. The number of cisternae in a stack can vary from 4-8, as typically seen in mammalian cells, to more than 30 in scale-secreting algae. The rims of the cisternae are often dilated and extended into tubules or tubular networks. Cisternal membranes are smooth from the lack of ribosomes and are usually curved, sometimes to the point of forming a circle rather than an arc. Gaps along the length of a cisterna are typical and represent holes, or fenestrae, within the disk. Different types of vesicles appear to bud from cisternal rims and carry proteins to other organelles (ER, plasma membrane, endosomes or lysosomes) or to other cisternae within the Golgi apparatus.
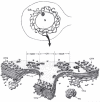
Depending on the cell type, an individual cell section may contain many Golgi stacks that appear to be separate structures, but this often represents a single Golgi ribbon winding in and out of the section plane. This can be seen in the electron micrograph of epithelial cells impregnated with osmium shown in (Fig. 1), which appears to show multiple Golgi stacks per cell. However, the inset shows a lateral view of a comparable sample, visualized with a light microscope, where the “classically stained” Golgi apparatus shows its ribbon-like character as it twists and turns through the apical portion of the cell.17 In addition, adjacent stacks of cisternae are often separated by what appear to be clusters of vesicles in thin sections, but when viewed in thick sections are actually tubular connections, or noncompact regions, between cisternae at equivalent positions in the stacks (labeled NCR in Fig. 2).18 Thus, it is generally thought, although difficult to prove, that the Golgi apparatus is a single-copy organelle in most mammalian cells. However, other cell types, such as plant cells specialized for secretion, may have several hundred separate Golgi stacks per cell (called dictyosomes in the older literature).19
The distinct polarity of the Golgi stack was apparent from early EM studies. As seen in Figure 1, cisternae at one edge of the Golgi stack preferentially reduced osmium and were blackened. This osmiophilic side of the Golgi apparatus is now known as the cis, or forming, face of the organelle. It is often adjacent to ER exit sites and small vesicular-tubular clusters (VTCs),20 representing ER to Golgi transport intermediates, are found between the ER and cis Golgi. The collection of VTCs, which are thought to be formed by the coalescence of COPII vesicles budding from the ER, represent the ER-Golgi intermediate compartment, or ERGIC.21 Medial cisternae comprise the middle of the stack and trans cisternae compose the exit face of the Golgi apparatus. The trans cisternae can be preferentially stained for acid phosphatase and thiamine pyrophosphatase,22,23 demonstrating a distinct enzyme composition for the trans face. In fact, each region appears to have a distinct composition as indicated by the observation that Golgi markers for the cis, medial and trans regions can be separated by density-gradient centrifugation.24 The last one or two cisternae at the trans side tend to be more reticulated and often appear to be peeling off of the stack (Fig. 2). This part of the Golgi apparatus is called the trans-Golgi network (TGN)25 or trans-tubular network (TTN)26 and produces large secretory granules in specialized secretory cells. Clathrin-coated vesicles specifically bud from the TGN in all cell types and provide a morphological signpost for the TGN.27 It is important to note that the cis, medial, trans, and TGN labels apply to regions within a continuum of cisternae and it is usually impossible to know, for example, where the cis-Golgi ends and the medial Golgi begins.
The position of the Golgi within the cell varies with cell type and species. The Golgi apparatus will often sit between the nucleus and the apical membrane of polarized epithelial cells (Fig. 1), but takes a perinuclear position in nonpolarized mammalian cells (Fig. 3). This is controlled by the microtubule network of the cell and dynein/dynactin motor proteins that transport Golgi elements to the microtubule organizing center sitting adjacent to the nucleus.28 In plant cells, the Golgi complex appears to be randomly distributed throughout the cytoplasm. 19
Three-Dimensional (3D) Structure of the Golgi Apparatus
The heterogenous and convoluted structure of the Golgi apparatus presents a challenge to visualizing its 3D structure. One approach has been reconstruction from serial thin sections, in which EM images are collected from adjacent thin sections (<100 nm) of the same sample and used to reconstruct the whole. In addition, Rambourg and Clermont were the first to investigate the 3D structure of Golgi using a stereoscopic approach,29 where two photographs of the same thick section (150-200 nm) are taken at two different angles by EM and then viewed with a stereoscope. These studies contributed to the view that the Golgi is a single-copy organelle,18 but even with these techniques, controversies over the autonomy of each Golgi cisternae were not resolved. For example, is each cisternae a separate compartment, or are adjacent cisternae connected by tubular elements?
With new techniques developed in recent years, the 3D ultra-structure of Golgi apparatus is being obtained at higher resolution, which has helped to refine our understanding of the relationship between its structure and function.30-32 The traditional chemical fixation of samples is replaced with fixation by ultra-rapid freezing, followed by freeze substitution. This method enables the immobilization of all molecules in a cell within milliseconds, thus reducing fixation artifacts to a minimum. Another improvement is the use of dual-axis, high-voltage electron microscope (HVEM) tomography to analyze of a series of relatively thick sections (1 μm compared to less than 100 nm in common transmission EM). The sections are tilted from +60° to -60° and photographed every 1.5° to generate a tilt series. The specimen is then rotated 90° in the plane of the grid and a second tilt series is taken. Compared to reconstruction of serial thin sections, less information is lost since fewer sections are required and the dual axis tilt series provides a resolution approaching 5 nm.
Two types of cultured mammalian cells were initially examined by this method with similar results.30,31,33 The Golgi apparatus from both of these cells contained 7 cisternae and adjacent stacks of Golgi were connected by tubular bridges between the cisternae at equivalent levels. However, there did not appear to be tubular connections between adjacent cisternae in the cis-trans direction, supporting the view that each cisterna is an autonomous structure. However, later reconstructions of Golgi in cells stimulated to increase secretory protein load showed evidence of tubular connections between nonadjacent cisternae that could mediate flow of protein independently of vesicular transport.34,35 All cisternae in the reconstructions were fenestrated and large cisternal holes were aligned to form “wells” within the stack. These “wells” are probably regions active in protein transport since they are filled with free vesicles and budding profiles. Tubules with budding tips extended perpendicularly from the margins of both cis and trans cisternae. The tubules from cis-side cisternae reached into the ERGIC region, and tubules from trans-side cisternae reached into the TGN regions. These tubules appear to play a role in protein transport, either by further fragmentation into transport vesicles, or release of an entire tubule carrying proteins to the cell surface. All cisternae displayed coated buds that were mainly located at their margins and at the edge of holes. Only the trans-most cisterna displayed clathrin-coated buds, whereas the others only displayed non-clathrin coated buds. One reconstructed segment of a ribbon contained ˜2100 vesicles in the Golgi region, giving the impression of a tremendous flux of membrane by vesicular transport meachanisms.31 The trans cisternae were wrapped with a specialized ER membrane, which lacked ribosome association at the face adjacent to the Golgi cisternae. The reason for an association between the trans Golgi and the ER is unknown, although it is possible that lipid exchange might occur in these regions of contact.
Structure of the Golgi Apparatus in S. cerevisiae
The budding yeast Saccharomyces cerevisae deserves special mention because it has become an important system for studying the secretory pathway and it was originally thought to be an exception to the rule that all eukaryotes have a Golgi apparatus. This is because stacked cisternae are rarely seen in S. cerevisiae. Instead, the Golgi cisternae are dispersed throughout the cytoplasm and are primarily disk-like or tubular networks.36,37 By staining cells with reduced osmium and viewing thick sections, these small tubular networks were found throughout the cells and were considered to correspond to Golgi structures.37 The tubules form a meshwork with dilated nodules at the intersections that appear to concentrate secretory cargo and give rise to secretory vesicles (˜100 nm vesicles). These structures disappear in mutants that block protein transport from the ER to the Golgi apparatus,37 and expand into structures more similar to mammalian cisternae (Berkeley bodies) in mutants that block protein exit from the Golgi apparatus.38 Protein transport and modification studies indicate that yeast have functionally distinct cis, medial, trans and TGN compartments, just as in other cell types.39,40 These studies used a temperature-sensitive sec18 (NSF) mutant to define the steps in glycoprotein maturation requiring vesicle-mediated protein transport (Fig. 4) and strongly argue that multiple SNARE-dependent membrane fusion events are required for intra-Golgi transport. Immunofluorescent localization of Golgi markers suggests that there are approximately 5-10 cisternae for each functionally distinct compartment.41-43
Protein Composition of the Golgi Apparatus
The three primary functions of the Golgi apparatus are the transport, sorting and modification of both protein and lipid, and the protein composition of the organelle reflects these functions. It is estimated that up to 1000 proteins make up the mammalian Golgi apparatus, and about 200 of these have been identified so far.44 With the development of high-throughput tandem mass spectrometry (MS) and the increasing availability of full genome sequences, subcellular proteomics has allowed more and more Golgi components to be identified.45,46 These include proteins required for: 1) glycosylation of proteins and lipids; 2) proteolytic processing of hormones and neuropeptides; 3) protein transport and sorting; 4) lipid synthesis and modification; 5) translocation of ions, heavy metals or lipids across the membrane; 6) the structure or inheritance of the Golgi apparatus (Golgi matrix proteins); and 7) cytoskeleton association and regulation. Table 1 provides examples of membrane proteins found associated with the mammalian and yeast Golgi apparatus that often serve as markers for this organelle.
Table 1
Membrane proteins of the mammalian and yeast Golgi apparatus.
Posttranslational Modifications Catalyzed within the Golgi Apparatus
Production of Glycoconjugates
The Golgi apparatus is an assembly line for the production of glycoconjugates through the sequential action of glycosyltransferases that add a diverse set of carbohydrates to both proteins and lipids. These glycoconjugates are important for many biological and disease processes, and play a critical role in self-nonself recognition, both across and within species.47 For example, host-pathogen interactions are often mediated through binding to specific carbohydrates. In addition, glycoconjugates are a major component of sperm-egg recognition and help prevent cross-species fertilization. Glycoconjugates are also major transplant antigens with the ABO blood system being the best-known example. In fact, transgenic pigs are being developed that express human Golgi glycosyltransferases with the goal of harvesting pig organs for transplantation that will not be rejected by the human immune system.48,49
A single species will produce a large number of different glycoconjugates, from glycoproteins (mostly protein) to proteoglycans (mostly carbohydrate) to glycolipids, and requires a large number of glycosyltransferases for this task. Each glycosyltransferase is specific for the sugar it transfers (e.g., galactose vs. sialic acid), the linkage used to attach the sugar to a growing polymer (e.g., α1->6 vs. α1->2), and the substrate receiving the sugar (e.g., N- vs O-linked oligosaccharides). Different species express different sets of Golgi glycosyltransferases and thus produce different glycoconjugates. An entire textbook is required for a full description of glycoconjugate biosynthesis, and so we will primarily restrict this discussion to the generation of a “typical” N-linked oligosaccharide on glycoproteins from mammals and yeast.
The process of N-linked glycosylation of proteins starts in the ER by the addition of a N-acetylglucosamine2Mannose9Glucose3 (GlcNAc2Man9Glc3) oligosaccharide structure on asparagine (N) residues in the sequence Asn-X-Ser/Thr. This oligosaccharide is pre-assembled on dolichol, a long chain lipid, and transferred en bloc by oligosaccharyltransferase to a nascent polypeptide emerging from the translocon. Then, the three glucose residues and typically one mannose residue are removed in the ER to generate the Asn-GlcNAc2Man8 “high-mannose” glycoprotein that is exported to the Golgi apparatus for further processing.50 The structure of the dolichol-GlcNAc2Man9Glc3 donor and subsequent glycan processing events in the ER appears to be remarkably well conserved in all eukaryotes, in contrast to the modification events in the Golgi apparatus. O-glycosylation also appears to be initiated in the ER, but in this case by transfer of a monosaccharide from either a dolichol-linked (Dol-P-Man in yeast) or sugar nucleotide (UDP-GalNAc in mammals) donor to serine or threonine residues.
N-Glycan Processing in the Mammalian Golgi Apparatus
In mammalian cells, production of “complex” N-glycans is initiated within early cisternae of the Golgi apparatus by the trimming of several more mannose residues by mannosidase I and II to produce an Asn-GlcNAc2Man3 structure. The chain is then extended by the sequential addition of GlcNAc, galactose and sialic acid in medial to TGN cisternae (Fig. 4). Fucose can also be added to the first GlcNAc attached to the Asn.50 Not all N-glycans are processed to this complex form. In particular, N-glycans on lysosomal enzymes are not as extensively processed by the Golgi mannosidases, leaving them in the high mannose form, and instead they are modified with a phospho-GlcNAc on the 6 position of certain mannose residues. This modification occurs in the cis Golgi and the GlcNAc is removed in a later compartment to generate the mannose-6-phosphate moiety required for sorting these glycoproteins to the lysosome (Fig. 4).51 High mannose and complex N-glycans can be distinguished experimentally by their sensitivity to endoglycosidase H (Endo H). Endo H can cleave high mannose N-glycans on glycoproteins in transit through the ER and early Golgi, but they become Endo H resistant as they are trimmed of mannose in cis Golgi cisternae and modified with GlcNAc in the medial Golgi. It is noteworthy that due to the diversity of the modifying enzymes in different cell types, even within an individual, the mature N-glycan structure attached to proteins are extremely variable.
Specific glycosyltransferases catalyze the transfer of the sugars described above from sugar nucleotide donors (UDP-GlcNAc, UDP-Gal, GDP-fucose and CMP-sialic acid) to the growing oligosaccharide chain. For most reactions, this generates a nucleotide diphosphate, which is then cleaved to the monophosphate by a nucleotide diphosphatase. Antiporters in the Golgi membrane then exchange the nucleotide monophosphate for a fresh sugar nucleotide. In the cytosol, the monophosphates are converted to triphosphates and enter the pool used to form new sugar nucleotides.52 This one-for-one exchange mediated by the antiporters ensures the availability of sugar nucleotide donors “on demand” in the Golgi lumen, without a wasteful accumulation of this energetically expensive precursor.
N-Glycan Processing in the Yeast Golgi Apparatus
In yeast, the process of N-glycosylation in the ER is the same as described above. However, complex N-glycans are not produced in the Golgi apparatus and yeast glycoproteins can be classified as “high mannose” and “extremely high mannose”. This is because the yeast Golgi apparatus lacks α-mannosidases and contains several different mannosyltransferases that extend the N-glycans with mannose. Glycoproteins destined for intracellular organelles receive a limited number of mannose residues (˜5 per N-glycan) in the Golgi apparatus, while many secreted glycoproteins are modified with 25 to more than 100 mannose residues to generate mannoproteins, an important component of the cell wall. This apparent simplicity in sugar content belies the large number of mannosyltransferases required to produce these glycoproteins (see Table 1). Mannose is added sequentially in three different linkages, α1->6, α1->2 and α1->3, in cis, medial, and trans cisternae, respectively (Fig. 4). The extent of mannose addition is determined by whether a single α1->6-mannose, or a long chain of α1->6- mannose is added to the N-glycan.53 Intermediates in this biosynthetic pathway can be identified using linkage-specific antibodies to the oligosaccharides and specific glycosidases. These reagents have been extremely useful for monitoring the progression of newly synthesized glycoproteins through the Golgi apparatus.39
Proteolytic Processing
A large number of secreted proteins, such as serum albumin, insulin, glucagon and many other peptide hormones, are initially synthesized as high molecular weight precursors called proproteins. Proteolytic processing of the proprotein is initiated by cleavage at dibasic sites (Arg-Arg, Arg-Lys or Lys-Lys) within the TGN or secretory granules formed from the TGN. A family of subtilisin-like proteases responsible for this processing event includes furin and PC1 - PC7 (prohormone convertase) from mammals and Kex2 from yeast. These endoproteases often work in concert with carboxypeptidases and/or aminopeptidases to process proproteins to their biologically active mature form.54-56
In recent years, the Brown and Goldstein lab has discovered another set of Golgi proteases involved in processing a high molecular weight, membrane-bound precursor of the sterol regulatory element binding protein (SREBP), a transcription factor that regulates expression of cholesterol and fatty acid biosynthetic genes.57 The SREBP precursor spans the membrane twice, with the N-terminal transcription factor and C-terminal regulatory domains facing the cytosol. When cholesterol levels in the ER membrane drop, SREBP is transported to the Golgi apparatus where it encounters site-1 protease (S1P) and site-2 protease (S2P). S1P cleaves the lumenal loop of SREBP separating the two halves of this protein, and S2P releases the N-terminal transcription factor domain by cleaving within the second transmembrane domain. This unusual proteolytic activity of S2P, occurring within the hydrophobic confines of the membrane bilayer, is shared by the presenilin-dependent γ-secretase, another Golgi-associated protease that produces the amyloid β peptide thought to cause Alzheimer's disease.58 Therefore, regulated intramembrane proteolyis, or RIP, within the Golgi apparatus plays a critically important role in cardiovascular and mental health.57
Protein Transport and Sorting in the Golgi Apparatus
General Mechanisms and Pathways
In eukaryotes, different cellular functions are confined to specific membrane-bound organelles. Enzymes that mediate these functions are synthesized on cytosolic ribosomes (with the exception of proteins encoded by mitochondrial and chloroplast genomes) and thus need to be sorted and delivered from this site to the appropriate organelle. As originally described by Blobel,59 non-cytosolic proteins must contain a signal, or address label, that tells the cell where to put them. Other proteins (receptors) act as postmen reading the address labels by molecular recognition and delivering their protein cargo to their home organelle, or a “delivery truck” (transport vesicles or tubules) heading in the right direction. The Golgi apparatus is the sorting, packaging and distribution center of the exocytic pathway, handling proteins and lipids destined for the ER, plasma membrane, endosomes and lysosomes or the Golgi itself (Fig. 5).
Membrane-bound vesicles, often wearing a proteinacious coat, mediate protein transport from the Golgi apparatus to other organelles.60-62 These vesicle coat proteins are thought to bend the membrane during vesicle budding, and also help to select and concentrate cargo proteins within the vesicles. Thus, the coat components often define the identity of these vesicles. A few types of coated vesicles generated from the Golgi apparatus have been well characterized that mediate different steps of protein transport. COPI-coated vesicles bud from all levels of the Golgi and are required for the retrograde transport of escaped ER residents back to the ER. These vesicles also appear to mediate protein transport between Golgi cisternae, although whether they mediate anterograde (forward) transport, retrograde transport, or both has been the subject of intense debate. Clathrin-coated vesicles (CCVs) form at the TGN, or immature secretory granules (ISG in Fig. 5), and carry proteins to endosomes. For example, lysosomal enzymes bearing the mannose-6-phosphate (M6P) determinant bind to the M6P receptor in the TGN and the complex is packaged into CCVs for initial delivery to an endosome. The lysosomal enzymes dissociate from the M6P receptor in the acidic environment of the endosome, allowing the receptor to recycle back to the TGN and the subsequent delivery of enzymes to the lysosome.63
In addition, the TGN produces several different secretory vesicles with no known coat. Some cells will produce both “constitutive” vesicles (or tubules) that will fuse to the plasma membrane without the need for a stimulus, as well as “regulated” vesicles (or secretory granules) that require a calcium influx to drive fusion with the plasma membrane (Chapter 5). Moreover, polarized cells will segregate apical from basolateral proteins in the TGN and package these proteins into distinct vesicles.64 The basolateral proteins appear to be packaged into CCVs at the TGN and whether these vesicles are targeted directly to the basolateral membrane or initially to an endosome is unclear.65,66 Mechanisms for transport vesicle formation and targeting will be covered in greater detail in other chapters.
Golgi Protein Localization
Golgi resident proteins, such as the glycosyltransferases, are preferentially localized in different Golgi regions but the mechanism for localizing these enzymes to specific cisternae is unknown. All known Golgi glycosyltransferases are type II integral membrane proteins and localization signals have been found within the cytosolic tails, transmembrane domains (TMDs) and lumenal domains for different enzymes.67 No specific sequence comprising a Golgi localization signal that is shared by multiple proteins, such as the KDEL motif found on soluble ER proteins,68 has been identified thus far. Nor has any “receptor” been defined that recognizes a Golgi localization signal operating in the cis - trans cisterna. The situation is a little better for TGN resident proteins such as furin and Kex2p, where specific signals in the cytosolic tails mediate localization.55,69 These proteins appear to cycle to endosomes, and/or the plasma membrane as part of their normal trafficking itinerary. The cytosolic tail signals of these proteins are similar to endocytosis signals and operate in retrieval from endosomes back to the TGN.55 The rest of this discussion will focus on proteins localized in cis - trans cisternae.
For many years, it was assumed that Golgi cisternae were stable structures and that resident enzymes were statically retained within a cisterna. Models for Golgi protein localization that were popular in the 1990's reflected this bias. For example, the “oligomerization” or “kin recognition” hypothesis suggested that residents of a particular cisterna (kin) would form aggregates that were too large to enter into vesicles moving cargo in the anterograde direction, and thus these aggregates were retained in the Golgi cisterna in which they were formed.70,71 While there is evidence for interaction between Golgi proteins,72 they do not appear to form large oligomers in vivo.73 In addition, the Golgi apparatus appears to transport large oligomers, such as collagen or algal scales, through the stack fairly efficiently.74,75 Therefore, formation of large oligomers per se would not prevent movement through the Golgi, and the “kin recognition” hypothesis, at least as originally proposed, appears to be untenable.
A second “bilayer-thickness” hypothesis stemmed from the observation that the length of a TMD Golgi localization signal seemed more important than its amino acid sequence.76 Bretscher and Munro noted that Golgi enzymes tend to have shorter TMDs than plasma membrane proteins. They suggested that differences in membrane thickness across the Golgi stack, determined by differences in cholesterol content, caused Golgi enzymes to partition into membranes with an appropriate bilayer thickness to fit the length of their TMDs.77 This partitioning would prevent the lateral diffusion of Golgi enzymes into forming anterograde vesicles with thicker bilayers. However, studies in insect cells indicate that cholesterol is not a major determinant of Golgi protein localization,78 and whether or not bilayer thickness, controlled by another means, contributes to this process has not been experimentally tested. Moreover, it appears that bilayer thickness is determined primarily by the protein component rather than the lipid component of membranes.79 Therefore, the high concentration of Golgi enzymes likely determines the thickness of the bilayer in Golgi membranes, rather than cholesterol content, and perhaps this serves a mechanism to reinforce segregation of Golgi enzymes from non-Golgi membrane proteins. It should also be noted that the bilayer thickness model suggests a mechanism for how localization signals within the transmembrane segment function, but does not explain how localization signals in the cytosolic tails and lumenal domains of different Golgi proteins operate.
Studies on the localization of two different Golgi glycosyltransferases from yeast and one from mammalian cells suggested that these proteins were not significantly “retained” in their compartment of residence, but were actively retrieved from later Golgi compartments.80-82 These observations suggested a more dynamic mechanism for Golgi protein localization than previously considered, analogous to the KDEL-dependent retrieval of ER proteins from the Golgi apparatus back to the ER in COPI-coated transport vesicles.83 In fact, COPI-coated vesicles appear to mediate retrograde transport of Golgi enzymes back to the ER84 and from later to earlier Golgi cisternae.85-87 At least some Golgi proteins continuously cycle all the way back into the ER as part of their normal trafficking itinerary, and it has been argued that all Golgi proteins continuously transit through the ER.84,88-90 Thus, while the mechanism for Golgi protein recognition (i.e., a sorting receptor) is not defined, it appears that retrograde transport plays an important role in Golgi protein localization. The growing realization that Golgi enzymes are not static residents of cisternae has impacted current views on how proteins move through the Golgi in the anterograde direction.
Protein Transport Through the Golgi Apparatus
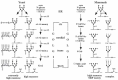
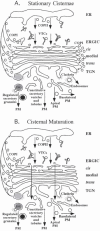
The mechanism by which secretory cargo moves through the Golgi apparatus is unknown although two very different models have been proposed and hotly debated. The “stationary cisternae / vesicular transport” model suggests that each cisterna of the Golgi is a stable entity and secretory cargo is transported from one cisterna to the next in vesicles moving in the anterograde direction. Proteins enter the Golgi by fusion of VTCs with a preexisting cis-most cisterna and exit from the TGN by being packaged into larger secretory vesicles for delivery to the plasma membrane (Fig. 5A). The alternative “cisternal maturation” (or cisternal progression) model suggests that the cis cisterna forms de novo by fusion of ER-derived membrane (VTCs) and progressively moves down the stack towards the trans face as though on a conveyer belt, maturing into medial and then trans cisternae along the way. The TGN then fragments into vesicles and is thus consumed (Fig. 5B). Cisternae are thought to mature by exporting cis Golgi enzymes in transport vesicles (COPI-coated) to a younger cisterna forming in the rear, while acquiring later Golgi enzymes from older cisternae. The latter model suggests that the Golgi is an outgrowth of the ER and this is consistent with the effect of brefeldin A on the Golgi. This drug induces a collapse of early Golgi cisternae into the ER and later compartments with endosomes, but after the drug is removed, Golgi enzymes are exported from the ER and the stack is rebuilt.91,92 Remarkably, the entire organelle can be disassembled by brefeldin A and rebuilt within a few minutes after the drug is removed, indicating a tremendous plasticity for this organelle.
The history of the two models for protein transport through the Golgi is quite interesting. Electron microscopists studying the Golgi apparatus in the late 1950's and 1960's originally suggested that cisterna are produced on the “forming face”, progress across the stack and are consumed into secretory vesicles at the “maturing face”.13,93-96 However, as various Golgi markers became better characterized, it was argued that cisternal progression couldn't adequately explain how resident proteins stay in the Golgi apparatus as secretory material quickly passes through. Nor did it explain how the Golgi residents could be concentrated in specific cisternae or the role of the numerous small vesicles surrounding the stack.15 Moreover, secretory cargo seemed to move quite efficiently between two different Golgi stacks in experimentally fused cells.97 Thus, in the 1980s several investigators suggested that a stable compartment model with secretory material passing from cisterna to cisterna in vesicles would better explain the available data.15,24,98,99 This model was boosted by the reconstitution of vesicle-mediated protein transport between Golgi cisternae by the Rothman lab,100 which provided a tremendous advance in defining the molecular mechanisms of protein transport. This in vitro assay was designed to measure the movement of VSV G protein (secretory cargo) in transport vesicles from purified “donor” Golgi membranes deficient for GlcNAc transferase to “acceptor” Golgi membranes containing this enzyme but lacking VSV G. It led to the discovery of coatomer (COPI),101 the role of the small GTP-binding protein ARF in budding COPI vesicles,102 and the function of NSF and SNARE proteins in vesicle targeting and fusion.103-105
While the stationary cisternae/vesicular transport model enjoyed substantial popularity in the 1980's and most of the 1990's, it was not universally accepted.106 Morphologists studying secretion of scales from algae continued to make a particularly good case for cisternal progression. 74 These carbohydrate rich structures are large enough to be visible in electron micrographs, and in many species the scales are significantly larger than the COPI vesicles surrounding the Golgi apparatus. A wave of scale secretion can be induced by deflagellating the algae, and the scales are observed to move across the Golgi stack without entering into small transport vesicles.107 This mode of transport does not appear to be unique to algae as other groups has made similar observations for the secretion of collagen from mammalian fibroblasts.75,108 Collagen is a 300 nm long, rod-shaped protein that forms large electron-dense aggregates within the Golgi apparatus. Its folding into a triple helical conformation within the ER requires an unusual hydroxyproline modification and the iron-dependent prolyl hydroxylase can be reversibly inhibited by iron chelators. Thus, cells treated with the chelator accumulate unfolded procollagen in the ER and a wave of collagen secretion can be induced by removing the chelator. In these experiments, collagen was observed to travel across the stack of Golgi cisternae without entering into smaller vesicles.75 However, this interpretation is partly based on the assumption that anterograde vesicles are constrained to a 50-60 nm diameter. In similar studies, the Rothman and Orci groups argued that a different protein aggregate in transit through the Golgi could be found in large “megavesicles” adjacent to cisternae.109 While these studies did not resolve the controversy, the pendulum of consensus view started swinging back toward the cisternal maturation model.110-112
The role of COPI-coated vesicles is another important distinguishing feature of these two models and the discovery that COPI vesicles mediate retrograde transport to the ER further swayed opinion towards the cisternal maturation model. In the stationary cisternae model, COPI vesicles are proposed to carry cargo in the forward (anterograde) direction between cisternae, whereas with the cisternal maturation model, these vesicles are proposed to carry resident Golgi enzymes in the retrograde direction. COPI was first implicated in retrograde protein transport from a genetic screen in yeast for mutants defective in Golgi to ER retrograde transport of a reporter protein bearing a “KKXX” ER retrieval signal. Mutant alleles for most of the COPI subunits were isolated in this screen, which demanded a functional secretory pathway for delivery of the KKXX-reporter protein to the plasma membrane.113 COPI mutants isolated in this and other unbiased screens38,114 all exhibit a defect in retrograde transport but transport many proteins efficiently through the Golgi to the cell surface at the nonpermissive temperature. Some proteins are trapped in the ER of COPI mutants and this is thought to reflect a defect in recycling cargo receptors needed for packaging these proteins in COPII vesicles.114 The role of COPI in mediating Golgi to ER retrograde transport is now well established, 61,62 but remember that COPI was initially discovered using an in vitro assay that reconstituted vesicle-mediated transport between Golgi cisternae.101 However, this assay may reconstitute packaging of the GlcNAc transferase into COPI vesicles and the delivery of this modifying enzyme to Golgi cisternae containing VSV G.86,87 These findings suggest that COPI vesicles can also mediate retrograde transport of resident Golgi enzymes between Golgi cisternae.
Immunoelectron microscopy has also been used to probe the contents of peri-Golgi COPI vesicles with conflicting reports.115 One group reported finding significant levels of Golgi resident enzymes and the KDEL-receptor in these vesicles while VSV G was largely absent.85 Another group reported finding two populations of COPI vesicles, one containing retrograde cargo (the KDEL-receptor) and the other containing VSV-G.116 The latter observation led to a hybrid model postulating that cisternal maturation was a “slow track” through the Golgi apparatus while smaller cargo could speed through the stack using the COPI vesicle “fast track”.117 However, a direct comparison of the rate of transport for collagen and VSV G suggests that these two proteins move synchronously through the stack.118 Discrepancies in reports of COPI vesicle content and function may be explained by the growing evidence for discrete subpopulations of these vesicles. The Golgi region contains a number of large, coiled coil protein complexes called golgins that can tether vesicles to Golgi cisternae and perhaps control movement of vesicles across the stack. The CASP/golgin-84 complex can specifically bind to COPI vesicles containing Golgi enzymes but lacking ER retrograde or anterograde markers, whereas the p115-golgin tether can select COPI vesicles containing an anterograde cargo but lacking Golgi enzymes.119 Thus, it appears that not all COPI vesicles are created equal and perhaps each distinct transport step between Golgi cisternae uses a specific subpopulation of COPI vesicles.
However, the prevailing view that COPI vesicles are the major mediator of protein flux through the Golgi (anterograde or retrograde) may be inaccurate, and therefore the premise of using COPI vesicle content to distinguish competing models may not rest on a firm foundation. Yeast genetic studies indicate that anterograde transport of secretory cargo through the Golgi apparatus is efficient in the absence of COPI function,113,114 while inactivation of Sec18 (NSF), and thereby SNARE function, immediately blocks anterograde movement of cargo within the Golgi.120,121 This suggests that anterograde transport requires multiple membrane fusion events that are independent of a COPI vesicle intermediate. Some Golgi proteins are mislocalized to a downstream compartment (the vacuole) in the COPI mutants,84,122 notably those that cycle back to the ER, although the Golgi retains sufficient enzyme content to terminally glycosylate secretory cargo.123 If COPI is the sole mediator of retrograde transport of Golgi enzymes during cisternal maturation, we would expect a wholesale loss of the Golgi enzymes after inactivating COPI, which does not seem to occur. Moreover, there is good evidence for COPI-independent retrograde transport of Golgi enzymes to the ER in mammalian cells via a mechanism requiring Rab6 but no known vesicle coat protein. What mediates protein flux through this organelle if not COPI? One possibility is that transient tubular connections between unlike cisternae provide a conduit for the flow of Golgi enzymes in the retrograde direction to drive cisternal maturation, or anterograde flow of cargo through stable cisternae.34,35 These tubular connections would presumably require the SNARE machinery to form without a need for coat proteins, but how the directionality of protein flow would be controlled by this mechanism is unclear. Another possibility is that transport is mediated by an undefined vesicular intermediate that does not require COPI for formation.
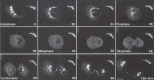
Perhaps the most direct test of the cisternal maturation model is to visualize individual cisternae in living cells over time to determine if the content of resident enzymes changes or stays the same. Because of their close proximity to each other, individual cisternae of the mammalian Golgi cannot be distinguished by light microscopy, but the scattered cisternae of Saccharomyces cerevisiae are ideal for this type of analysis. Two different groups have used GFP and RFP fused to different Golgi proteins (markers of cis, medial and trans compartments) to monitor the residence time of these markers in individual cisterna.124,125 By the stationary cisternae model, one would expect a relatively long residence time for these proteins within their cisternae. Instead, cis-Golgi cisternae marked with a GFP fusion protein rapidly lost their green color while they acquired the red color of a trans-Golgi protein fused to RFP. Importantly, the rate of this color change was very similar to the rate of anterograde cargo transport through the Golgi. In addition, the color changes were always unidirectional in the cis to trans direction; a trans cisterna never acquired cis-Golgi enzymes. These data are inconsistent with a stationary cisternae model and strongly support a cisternal maturation model. Interestingly, Golgi cisternae matured in a COPI mutant although the rate of maturation was slower relative to a wild-type cell.124
The majority of evidence in the literature now favors the cisternal maturation model and most investigators in the field have returned to this original view of a dynamic organelle in constant flux.110,112,126 However, some observations are difficult to reconcile with cisternal maturation. For example, incubation of mammalian cells at 20°C blocks exit of secreted proteins from the TGN while transport from the ER to the Golgi and through the stack is not inhibited. By the cisternal maturation model, one might anticipate that the 20°C block would cause an increase in the number of Golgi cisterna, but this does not occur. Instead cargo accumulates in bulging domains in the last few cisternae.32 To be consistent with a cisternal maturation mechanism, the TGN cisternae that are not consumed by fragmentation at 20C would have to join together by a homotypic fusion mechanism, which would suggest the number of cisternae in the cell is somehow tightly regulated.
Inheritance of the Golgi Apparatus
As a single-copy organelle, the Golgi apparatus must be partitioned during the process of cell division to ensure both daughter cells inherit a functional Golgi.127 The strategy for doing this appears to vary in different organisms. Some single-celled organisms divide the Golgi stack down the middle and separate the two halves to daughter cells.128,129 In mammalian cells, the Golgi apparatus undergoes a massive disassembly process during mitotic division, which is accompanied by a block in protein transport through the secretory pathway. This process is initiated in prophase with the fragmentation of the Golgi ribbon into multiple stacks that distribute around the nucleus in association with the mitotic spindle. A second stage of disassembly occurs in prometaphase when intrinsic Golgi proteins become finely dispersed throughout the cytoplasm (Fig. 6).130 The fate of Golgi proteins and membranes during this second stage has been a subject of controversy. Some investigators argue that all Golgi membranes are completely absorbed back into the ER and are then reassembled de novo starting in telophase.131 Others have presented evidence that the Golgi breaks down into numerous small vesicles and clustered Golgi fragments that remain separate from the ER.132-134 In this case, reassembly would only require fusion of vesicles and fragments derived from the same cisternae. Peripherally associated Golgi matrix proteins (such as GM130 and GRASP65) also undergo the initial fragmentation in prophase and remain associated with Golgi stacks, but these proteins do not appear to disperse throughout the cytosol in prometaphase. In addition, these matrix fragments can be segregated normally during mitosis in cells treated with brefeldin A. This suggests that the Golgi matrix can be segregated independently of Golgi membranes and may be the partitioning unit of inheritance, which then serves to nucleate Golgi reassembly adjacent to daughter cell nuclei during telophase.135 Interestingly, injection of a GRASP65 peptide into cells inhibits the initial fragmentation in vivo and blocks mitosis. However, these cells will enter mitosis when also treated with brefeldin A to disrupt the Golgi, suggesting that Golgi fragmentation is an essential prelude to mitosis.136
In vitro assays using semi-intact cells or purified Golgi stacks have been used to probe the mechanism of Golgi fragmentation. The initial stage of Golgi disassembly appears to be regulated by polo-like and MEK1 kinases,136,137 although the substrates relevant to Golgi breakdown are not yet known. The second stage of Golgi disassembly is thought to occur by the budding of COPI vesicles from cisternal rims and inhibition of their subsequent fusion (NSF-dependent heterotypic fusion) with target membranes.138 The central core of each cisterna then fragments in a COPI-independent fashion that may result from inhibition of homotypic membrane fusion driven by the NSF-like ATPase p97.139 Normal fissioning of the cisternae (by an unknown mechanism) would then fail to be balanced by fusion and lead to fragmentation. These events are controlled, at least in vitro, by the cyclin-dependent kinase CDK1.140,141 In telophase, when CDK1 activity drops, the Golgi vesicles and fragments cluster together and fuse to regenerate the Golgi apparatus in the new cells. The reassembly process seems to be driven by SNARE-dependent membrane fusion requiring both NSF and p97 ATPases.127
Summary
This chapter has emphasized the numerous issues surrounding the Golgi apparatus that are unresolved. These include a basic understanding of the relationship between form and function, how the Golgi is assembled and inherited, and how proteins move through this organelle. The pendulum has swung back to cisternal maturation as the most popular model to describe anterograde transport of secretory proteins through this organelle. However, many questions remain concerning the mechanism of cisternal maturation. For example, what is the precise role of COPI-coated vesicles in the maturation process. What is the contribution of transient inter-cisternal tubular connections? Do these direct inter-cisternal connections represent a COPI-independent mode of retrograde transport or are there undiscovered classes of transport vesicles that contribute to cisternal maturation? What governs the trafficking of resident Golgi enzymes and determines their steady-state localization to different cisternae in the stack? To what extent does cisternal maturation rely on cycling of Golgi enzymes through the ER relative to retrograde transport back one step to a younger cisterna? What triggers fragmentation of the TGN into multiple transport carriers with distinct cargos? Is clathrin and clathrin adaptors the only coat proteins that drives this protein sorting and fragmentation process or are other undiscovered coat proteins involved? Young investigators entering this field should find ample opportunity for making new discoveries that will help answer some of these questions.
References
- 1.
- Golgi C. Sur le structure des cellules nerveuses. Arch Ital Biol. 1898;30:60–71.
- 2.
- Cajal SR. Algunas variaciones fisiologicas y patologicas del aparato reticular de Golgi. Trab Lab Inv Biol Madr. 1914;12:127–227.
- 3.
- Negri A. Di una fina particolarita de struttura delle cellule di alcune ghiandole dei mammiferi. Boll Soc med-chir di Pavia. 1900;13-14:69–71.
- 4.
- Bentivoglio M. The Golgi apparatus emerges from nerve cells. Trends Neurosci. 1998;21(5):195–200. [PubMed: 9610881]
- 5.
- Fuchs H. Uber das epithel im nebenhoden der maus. Anat Hefte. 1902;19:313–347.
- 6.
- Nassonov DN. Das Golgische binnennetz und seine beziehungen zu der sekretion: Untersuchungen uber einige Amphibiendrusen. Arch Mikrosk Anat. 1923;97:136–186.
- 7.
- Bowen RH. The cytology of glandular secretion. Quart Rev Biol. 1929;4(299-324):484–519.
- 8.
- Baker JR. What is the Golgi controversy? J Roy Micr Soc. 1955;74:217–221. [PubMed: 14368608]
- 9.
- Dalton AJ, Felix MD. Cytologic and cytochemical characteristics of the Golgi substance of epithelial cells of the epididymis in situ, in homogenates and after isolation. Am J Anat. 1954;94(2):171–207. [PubMed: 13148119]
- 10.
- Dalton AJ, Felix MD. A comparative study of the Golgi complex. J Biophys Biochem Cytol. 1956;2(4 Suppl):79–84. [PMC free article: PMC2229724] [PubMed: 13357526]
- 11.
- Farquhar MG, Rinehart JF. Endocrin. 1954. pp. 857–876. [PubMed: 13210333]
- 12.
- Sjostrand FS, Hanzon V. Ultrastructure of Golgi apparatus of exocrine cells of mouse pancreas. Exp Cell Res. 1954;7:415–429. [PubMed: 13220587]
- 13.
- Beams HW, Kessel RG. The Golgi apparatus: Structure and function. Int Rev Cytol. 1968;23:209–276. [PubMed: 4873473]
- 14.
- Berger EG. The Golgi apparatus: From discovery to contemporary studies. In: Roth J, ed. The Golgi Apparatus. Basel, Boston and Berlin: Birkhauser Verlag. 1997:37–62.
- 15.
- Farquhar MG, Palade GE. The Golgi apparatus (complex)-(1954-1981)-from artifact to center stage. J Cell Biol. 1981;91(3 Pt 2):77s–103s. [PMC free article: PMC2112780] [PubMed: 7033246]
- 16.
- Whaley WG. The Golgi apparatus. Vienna and New York: Springer-Verlag. 1975
- 17.
- Friend DS, Murray MJ. Osmium impregnation of the Golgi apparatus. Am J Anat. 1965;117:135–149. [PubMed: 14345830]
- 18.
- Rambourg A, Clermont Y, Hermo L. Three-dimensional architecture of the Golgi apparatus in Sertoli cells of the rat. Am J Anat. 1979;154:455–476. [PubMed: 86291]
- 19.
- Dupree P, Sherrier DJ. The plant Golgi apparatus. Biochim Biophys Acta. 1998;1404(1-2):259–270. [PubMed: 9714825]
- 20.
- Balch WE, McCaffery JM, Plutner H. et al. Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell. 1994;76(5):841–852. [PubMed: 8124720]
- 21.
- Hauri HP, Schweizer A. The endoplasmic reticulum-Golgi intermediate compartment. Curr Opin Cell Biol. 1992;4(4):600–608. [PMC free article: PMC7134740] [PubMed: 1419041]
- 22.
- Novikoff PM, Novikoff AB, Quintana N. et al. Golgi apparatus, GERL, and lysosomes of neurons in rat dorsal root ganglia, studied by thick section and thin section cytochemistry. J Cell Biol. 1971;50(3):859–886. [PMC free article: PMC2108306] [PubMed: 4329159]
- 23.
- Goldfischer S, Essner E, Novikoff AB. The localization of phosphatase activities at the level of ultrastructure. J Histochem Cytochem. 1964;12:72–95. [PubMed: 14187314]
- 24.
- Dunphy WG, Rothman JE. Compartmental organization of the Golgi stack. Cell. 1985;42(1):13–21. [PubMed: 3926324]
- 25.
- Griffiths G, Simons K. The trans Golgi network: Sorting at the exit site of the Golgi complex. Science. 1986;234(4775):438–443. [PubMed: 2945253]
- 26.
- Rambourg A, Clermont Y. Three-dimensional structure of the Golgi apparatus in mammalian cells. In: Roth J, ed. The Golgi apparatus. Basel, Boston and Berlin: Birkhauser Verlag. 1997:1–36.
- 27.
- Pearse BM, Robinson MS. Clathrin, adaptors, and sorting. Annu Rev Cell Biol. 1990;6:151–171. [PubMed: 2177341]
- 28.
- Rios RM, Bornens M. The Golgi apparatus at the cell centre. Curr Opin Cell Biol. 2003;15(1):60–66. [PubMed: 12517705]
- 29.
- Rambourg A, Clermont Y, Marraud A. Three-dimensional structure of the osmium-impregnated Golgi apparatus as seen in the high voltage electron microscope. Am J Anat. 1974;140(1):27–45. [PubMed: 4132934]
- 30.
- Ladinsky MS, Mastronarde DN, McIntosh JR. et al. Golgi structure in three dimensions: Functional insights from the normal rat kidney cell. J Cell Biol. 1999;144(6):1135–1149. [PMC free article: PMC2150572] [PubMed: 10087259]
- 31.
- Marsh BJ, Mastronarde DN, Buttle KF. et al. Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc Natl Acad Sci USA. 2001;98(5):2399–2406. [PMC free article: PMC30150] [PubMed: 11226251]
- 32.
- Ladinsky MS, Wu CC, McIntosh S. et al. Structure of the Golgi and distribution of reporter molecules at 20 degrees C reveals the complexity of the exit compartments. Mol Biol Cell. 2002;13(8):2810–2825. [PMC free article: PMC117944] [PubMed: 12181348]
- 33.
- Marsh BJ, Mastronarde DN, McIntosh JR. et al. Structural evidence for multiple transport mechanisms through the Golgi in the pancreatic beta-cell line, HIT-T15. Biochem Soc Trans. 2001;29(Pt 4):461–467. [PubMed: 11498009]
- 34.
- Trucco A, Polishchuk RS, Martella O. et al. Secretory traffic triggers the formation of tubular continuities across Golgi sub-compartments. Nat Cell Biol. 2004;6(11):1071–1081. [PubMed: 15502824]
- 35.
- Marsh BJ, Volkmann N, McIntosh JR. et al. Direct continuities between cisternae at different levels of the Golgi complex in glucose-stimulated mouse islet beta cells. Proc Natl Acad Sci USA. 2004;101(15):5565–5570. [PMC free article: PMC397423] [PubMed: 15064406]
- 36.
- Preuss D, Mulholland J, Franzusoff A. et al. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell. 1992;3(7):789–803. [PMC free article: PMC275635] [PubMed: 1381247]
- 37.
- Rambourg A, Jackson CL, Clermont Y. Three dimensional configuration of the secretory pathway and segregation of secretion granules in the yeast Saccharomyces cerevisiae. J Cell Sci. 2001;114(Pt 12):2231–2239. [PubMed: 11493663]
- 38.
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. [PubMed: 6996832]
- 39.
- Brigance WT, Barlowe C, Graham TR. Organization of the yeast Golgi complex into at least four functionally distinct compartments. Mol Biol Cell. 2000;11(1):171–182. [PMC free article: PMC14766] [PubMed: 10637300]
- 40.
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114(2):207–218. [PMC free article: PMC2289075] [PubMed: 2071670]
- 41.
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113(3):527–538. [PMC free article: PMC2288974] [PubMed: 2016334]
- 42.
- Graham TR, Seeger M, Payne GS. et al. Clathrin-dependent localization of alpha 1,3 mannosyltransferase to the Golgi complex of Saccharomyces cerevisiae. J Cell Biol. 1994;127(3):667–678. [PMC free article: PMC2120240] [PubMed: 7962051]
- 43.
- Rayner JC, Munro S. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J Biol Chem. 1998;273(41):26836–26843. [PubMed: 9756928]
- 44.
- Taylor RS, Wu CC, Hays LG. et al. Proteomics of rat liver Golgi complex: Minor proteins are identified through sequential fractionation. Electrophoresis. 2000;21(16):3441–3459. [PubMed: 11079564]
- 45.
- Wu CC, Yates IIIrd JR, Neville MC. et al. Proteomic analysis of two functional states of the Golgi complex in mammary epithelial cells. Traffic. 2000;1(10):769–782. [PubMed: 11208067]
- 46.
- Bell AW, Ward MA, Blackstock WP. et al. Proteomics characterization of abundant Golgi membrane proteins. J Biol Chem. 2001;276(7):5152–5165. [PubMed: 11042173]
- 47.
- Varki A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology. 1993;3(2):97–130. [PMC free article: PMC7108619] [PubMed: 8490246]
- 48.
- Sharma A, Okabe J, Birch P. et al. Reduction in the level of Gal(alpha1,3)Gal in transgenic mice and pigs by the expression of an alpha(1,2)fucosyltransferase. Proc Natl Acad Sci USA. 1996;93(14):7190–7195. [PMC free article: PMC38958] [PubMed: 8692967]
- 49.
- Lai L, Kolber-Simonds D, Park KW. et al. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002;295(5557):1089–1092. [PubMed: 11778012]
- 50.
- Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291(5512):2364–2369. [PubMed: 11269317]
- 51.
- Goldberg DE, Kornfeld S. Evidence for extensive subcellular organization of asparagine-linked oligosaccharide processing and lysosomal enzyme phosphorylation. J Biol Chem. 1983;258(5):3159–3165. [PubMed: 6402509]
- 52.
- Berninsone PM, Hirschberg CB. Nucleotide sugar transporters of the Golgi apparatus. Curr Opin Struct Biol. 2000;10(5):542–547. [PubMed: 11042451]
- 53.
- Dean N. Asparagine-linked glycosylation in the yeast Golgi. Biochim Biophys Acta. 1999;1426(2):309–322. [PubMed: 9878803]
- 54.
- Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2(1):31–39. [PubMed: 9667917]
- 55.
- Thomas G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3(10):753–766. [PMC free article: PMC1964754] [PubMed: 12360192]
- 56.
- Fuller RS, Sterne RE, Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. [PubMed: 3288097]
- 57.
- Brown MS, Ye J, Rawson RB. et al. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100(4):391–398. [PubMed: 10693756]
- 58.
- Medina M, Dotti CG. RIPped out by presenilin-dependent gamma-secretase. Cell Signal. 2003;15(9):829–841. [PubMed: 12834808]
- 59.
- Blobel G. Protein targeting (Nobel lecture). Chembiochem. 2000;1(2):86–102. [PubMed: 11828402]
- 60.
- Bonifacino JS, Lippincott-Schwartz J. Coat proteins: Shaping membrane transport. Nat Rev Mol Cell Biol. 2003;4(5):409–414. [PubMed: 12728274]
- 61.
- Kirchhausen T. Three ways to make a vesicle. Nat Rev Mol Cell Biol. 2000;1(3):187–198. [PubMed: 11252894]
- 62.
- Springer S, Spang A, Schekman R. A primer on vesicle budding. Cell. 1999;97(2):145–148. [PubMed: 10219233]
- 63.
- Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: New twists in the tale. Nat Rev Mol Cell Biol. 2003;4(3):202–212. [PubMed: 12612639]
- 64.
- Traub LM, Kornfeld S. The trans-Golgi network: A late secretory sorting station. Curr Opin Cell Biol. 1997;9(4):527–533. [PubMed: 9261049]
- 65.
- Folsch H, Pypaert M, Maday S. et al. The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J Cell Biol. 2003;163(2):351–362. [PMC free article: PMC2173537] [PubMed: 14581457]
- 66.
- Folsch H, Ohno H, Bonifacino JS. et al. A novel clathrin adaptor complex mediates basolateral targeting in polarized epithelial cells. Cell. 1999;99(2):189–198. [PubMed: 10535737]
- 67.
- Opat AS, van Vliet C, Gleeson PA. Trafficking and localisation of resident Golgi glycosylation enzymes. Biochimie. 2001;83(8):763–773. [PubMed: 11530209]
- 68.
- Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48(5):899–907. [PubMed: 3545499]
- 69.
- Redding K, Seeger M, Payne GS. et al. The effects of clathrin inactivation on localization of Kex2 protease are independent of the TGN localization signal in the cytosolic tail of Kex2p. Mol Biol Cell. 1996;7(11):1667–1677. [PMC free article: PMC276017] [PubMed: 8930891]
- 70.
- Nilsson T, Slusarewicz P, Hoe MH. et al. Kin recognition: A model for the retention of Golgi enzymes. FEBS Letters. 1993;330:1–4. [PubMed: 8370450]
- 71.
- Weisz OA, Swift AM, Machamer CE. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J Cell Biol. 1993;122(6):1185–1196. [PMC free article: PMC2119850] [PubMed: 8397214]
- 72.
- Nilsson T, Hoe MH, Slusarewicz P. et al. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 1994;(13):562–574. [PMC free article: PMC394845] [PubMed: 8313901]
- 73.
- Cole NB, Smith CL, Sciaky N. et al. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 1996;273(5276):797–801. [PubMed: 8670420]
- 74.
- Becker B, Melkonian M. The secretory pathway of protists: Spatial and functional organization and evolution. Microbiol Rev. 1996;60(4):697–721. [PMC free article: PMC239460] [PubMed: 8987360]
- 75.
- Bonfanti L, Mironov Jr AA, Martinez-Menarguez JA. et al. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: Evidence for cisternal maturation. Cell. 1998;95(7):993–1003. [PubMed: 9875853]
- 76.
- Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. Embo J. 1995;14(19):4695–4704. [PMC free article: PMC394566] [PubMed: 7588599]
- 77.
- Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261(5126):1280–1281. [PubMed: 8362242]
- 78.
- Rolls MM, Marquardt MT, Kielian M. et al. Cholesterol-independent targeting of Golgi membrane proteins in insect cells. Mol Biol Cell. 1997;8(11):2111–2118. [PMC free article: PMC25695] [PubMed: 9362056]
- 79.
- Mitra K, Ubarretxena-Belandia I, Taguchi T. et al. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc Natl Acad Sci USA. 2004;101(12):4083–4088. [PMC free article: PMC384699] [PubMed: 15016920]
- 80.
- Graham TR, Krasnov VA. Sorting of yeast alpha 1,3 mannosyltransferase is mediated by a lumenal domain interaction, and a transmembrane domain signal that can confer clathrin-dependent Golgi localization to a secreted protein. Mol Biol Cell. 1995;6(7):809–824. [PMC free article: PMC301242] [PubMed: 7579696]
- 81.
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. Journal of Cell Biology. 1996;132(6):985–998. [PMC free article: PMC2120767] [PubMed: 8601597]
- 82.
- Hoe MH, Slusarewicz P, Misteli T. et al. Evidence for recycling of the resident medial/trans Golgi enzyme, N-acetylglucosaminyltransferase I, in ldlD cells. J Biol Chem. 1995;270(42):25057–25063. [PubMed: 7559636]
- 83.
- Pelham HR. Sorting and retrieval between the endoplasmic reticulum and Golgi apparatus. Curr Opin Cell Biol. 1995;7(4):530–535. [PubMed: 7495573]
- 84.
- Todorow Z, Spang A, Carmack E. et al. Active recycling of yeast Golgi mannosyltransferase complexes through the endoplasmic reticulum. Proc Natl Acad Sci USA. 2000;97(25):13643–13648. [PMC free article: PMC17629] [PubMed: 11095735]
- 85.
- Martinez-Menarguez JA, Prekeris R, Oorschot VM. et al. Peri-Golgi vesicles contain retrograde but not anterograde proteins consistent with the cisternal progression model of intra-Golgi transport. J Cell Biol. 2001;155(7):1213–1224. [PMC free article: PMC2199342] [PubMed: 11748250]
- 86.
- Love HD, Lin CC, Short CS. et al. Isolation of functional Golgi-derived vesicles with a possible role in retrograde transport. J Cell Biol. 1998;140(3):541–551. [PMC free article: PMC2140158] [PubMed: 9456315]
- 87.
- Lanoix J, Ouwendijk J, Lin CC. et al. GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COP I vesicles. Embo J. 1999;18(18):4935–4948. [PMC free article: PMC1171565] [PubMed: 10487746]
- 88.
- Wooding S, Pelham HR. The dynamics of golgi protein traffic visualized in living yeast cells. Mol Biol Cell. 1998;9(9):2667–2680. [PMC free article: PMC25539] [PubMed: 9725919]
- 89.
- Storrie B, White J, Rottger S. et al. Recycling of golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J Cell Biol. 1998;143(6):1505–1521. [PMC free article: PMC2132995] [PubMed: 9852147]
- 90.
- Cole NB, Ellenberg J, Song J. et al. Retrograde transport of Golgi-localized proteins to the ER. J Cell Biol. 1998;140(1):1–15. [PMC free article: PMC2132605] [PubMed: 9425149]
- 91.
- Lippincott-Schwartz J, Yuan L, Tipper C. et al. Brefeldin A's effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell. 1991;67(3):601–616. [PubMed: 1682055]
- 92.
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS. et al. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: Evidence for membrane cycling from Golgi to ER. Cell. 1989;56(5):801–813. [PMC free article: PMC7173269] [PubMed: 2647301]
- 93.
- Grasse PP. Ultrastructure, polarity and reproduction of Golgi apparatus. C R Hebd Seances Acad Sci. 1957;245(16):1278–1281. [PubMed: 13473197]
- 94.
- Grimstone AV. Fine structure and morphogenesis in Protozoa. Biol Rev Camb Philos Soc. 1961;36:97–150. [PubMed: 13708746]
- 95.
- Mollenhauer HH, Whaley WG. An observation on the functioning of the Golgi apparatus. J Cell Biol. 1963;17:222–225. [PMC free article: PMC2106267] [PubMed: 13935874]
- 96.
- Mollenhauer HH, Morre DJ. Golgi apparatus and plant secretion. Ann Rev Plant Physiol. 1966;17:27–46.
- 97.
- Rothman JE, Miller RL, Urbani LJ. Intercompartmental transport in the Golgi complex is a dissociative process: Facile transfer of membrane protein between two Golgi populations. J Cell Biol. 1984;99(1 Pt 1):260–271. [PMC free article: PMC2275621] [PubMed: 6539782]
- 98.
- Farquhar MG. Progress in unraveling pathways of Golgi traffic. Annu Rev Cell Biol. 1985;1:447–488. [PubMed: 3916320]
- 99.
- Rothman JE. The golgi apparatus: Two organelles in tandem. Science. 1981;213(4513):1212–1219. [PubMed: 7268428]
- 100.
- Balch WE, Dunphy WG, Braell WA. et al. Reconstitution of the transport of protein between successive compartments of the Golgi measured by the coupled incorporation of N-acetylglucosamine. Cell. 1984;39(2 Pt 1):405–416. [PubMed: 6498939]
- 101.
- Waters MG, Serafini T, Rothman JE. ‘Coatomer’: A cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature. 1991;349(6306):248–251. [PubMed: 1898986]
- 102.
- Serafini T, Orci L, Amherdt M. et al. ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: A novel role for a GTP-binding protein. Cell. 1991;67(2):239–253. [PubMed: 1680566]
- 103.
- Malhotra V, Orci L, Glick BS. et al. Role of an N-ethylmaleimide-sensitive transport component in promoting fusion of transport vesicles with cisternae of the Golgi stack. Cell. 1988;54(2):221–227. [PubMed: 3390865]
- 104.
- Weidman PJ, Melancon P, Block MR. et al. Binding of an N-ethylmaleimide-sensitive fusion protein to Golgi membranes requires both a soluble protein(s) and an integral membrane receptor. J Cell Biol. 1989;108(5):1589–1596. [PMC free article: PMC2115541] [PubMed: 2541136]
- 105.
- Sollner T, Whiteheart SW, Brunner M. et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362(6418):318–324. [PubMed: 8455717]
- 106.
- Mollenhauer HH, Morre DJ. Perspectives on Golgi apparatus form and function. J Electron Microsc Tech. 1991;17(1):2–14. [PubMed: 1993935]
- 107.
- McFadden GI, Melkonian M. Golgi apparatus activity and membrane flow during scale biogenesis in the green flagellate Scherffelia dubia (Prasinophyceae). I. Flagellar regeneration. Protoplasma. 1986;130(186-198)
- 108.
- Leblond CP. Synthesis and secretion of collagen by cells of connective tissue, bone, and dentin. Anat Rec. 1989;224(2):123–138. [PubMed: 2672880]
- 109.
- Volchuk A, Amherdt M, Ravazzola M. et al. Megavesicles implicated in the rapid transport of intracisternal aggregates across the Golgi stack. Cell. 2000;102(3):335–348. [PubMed: 10975524]
- 110.
- Allan BB, Balch WE. Protein sorting by directed maturation of Golgi compartments. Science. 1999;285(5424):63–66. [PubMed: 10390362]
- 111.
- Glick BS, Malhotra V. The curious status of the Golgi apparatus [comment] Cell. 1998;95(7):883–889. [PubMed: 9875843]
- 112.
- Pelham HR. Getting through the Golgi complex. Trends Cell Biol. 1998;8(1):45–49. [PubMed: 9695808]
- 113.
- Letourneur F, Gaynor EC, Hennecke S. et al. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79(7):1199–1207. [PubMed: 8001155]
- 114.
- Gaynor EC, Emr SD. COPI-independent anterograde transport: Cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136(4):789–802. [PMC free article: PMC2132489] [PubMed: 9049245]
- 115.
- Rabouille C, Klumperman J. Opinion: The maturing role of COPI vesicles in intra-Golgi transport. Nat Rev Mol Cell Biol. 2005;6(10):812–817. [PubMed: 16167055]
- 116.
- Orci L, Stamnes M, Ravazzola M. et al. Bidirectional transport by distinct populations of COPI-coated vesicles. Cell. 1997;90(2):335–349. [PubMed: 9244307]
- 117.
- Pelham HR, Rothman JE. The debate about transport in the Golgitwo sides of the same coin? Cell. 2000;102(6):713–719. [PubMed: 11030615]
- 118.
- Mironov AA, Beznoussenko GV, Nicoziani P. et al. Small cargo proteins and large aggregates can traverse the Golgi by a common mechanism without leaving the lumen of cisternae. J Cell Biol. 2001;155(7):1225–1238. [PMC free article: PMC2199327] [PubMed: 11756473]
- 119.
- Malsam J, Satoh A, Pelletier L. et al. Golgin tethers define subpopulations of COPI vesicles. Science. 2005;307(5712):1095–1098. [PubMed: 15718469]
- 120.
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a sec18(NSF) mutant. JCB. 1991;114:207–218. [PMC free article: PMC2289075] [PubMed: 2071670]
- 121.
- Brigance WT, Barlowe C, Graham TR. Organization of the yeast Golgi complex into at least four functionally distinct compartments. Mol Biol Cell. 2000;11(1):171–182. [PMC free article: PMC14766] [PubMed: 10637300]
- 122.
- Sato K, Sato M, Nakano A. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J Cell Biol. 2001;152(5):935–944. [PMC free article: PMC2198819] [PubMed: 11238450]
- 123.
- Gaynor EC, Emr SD. COPI-independent anterograde transport: Cargo-selective ER to Golgi protein transport in yeast COPI mutants. JCB. 1997;136(4):789–802. [PMC free article: PMC2132489] [PubMed: 9049245]
- 124.
- Matsuura-Tokita K, Takeuchi M, Ichihara A. et al. Live imaging of yeast Golgi cisternal maturation. Nature. 2006 [PubMed: 16699523]
- 125.
- Losev E, Reinke CA, Jellen J. et al. Golgi maturation visualized in living yeast. Nature. 2006 [PubMed: 16699524]
- 126.
- Glick BS, Malhotra V. The curious status of the Golgi apparatus. Cell. 1998;95(7):883–889. [PubMed: 9875843]
- 127.
- Shorter J, Warren G. Golgi architecture and inheritance. Annu Rev Cell Dev Biol. 2002;18:379–420. [PubMed: 12142281]
- 128.
- Benchimol M, Ribeiro KC, Mariante RM. et al. Structure and division of the Golgi complex in Trichomonas vaginalis and Tritrichomonas foetus. Eur J Cell Biol. 2001;80(9):593–607. [PubMed: 11675935]
- 129.
- Pelletier L, Stern CA, Pypaert M. et al. Golgi biogenesis in Toxoplasma gondii. Nature. 2002;418(6897):548–552. [PubMed: 12152082]
- 130.
- Colanzi A, Suetterlin C, Malhotra V. Cell-cycle-specific Golgi fragmentation: How and why? Curr Opin Cell Biol. 2003;15(4):462–467. [PubMed: 12892787]
- 131.
- Zaal KJ, Smith CL, Polishchuk RS. et al. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 1999;99(6):589–601. [PubMed: 10612395]
- 132.
- Shima DT, Haldar K, Pepperkok R. et al. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol. 1997;137(6):1211–1228. [PMC free article: PMC2132532] [PubMed: 9182657]
- 133.
- Jokitalo E, Cabrera-Poch N, Warren G. et al. Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J Cell Biol. 2001;154(2):317–330. [PMC free article: PMC2150754] [PubMed: 11470821]
- 134.
- Jesch SA, Linstedt AD. The Golgi and endoplasmic reticulum remain independent during mitosis in HeLa cells. Mol Biol Cell. 1998;9(3):623–635. [PMC free article: PMC25291] [PubMed: 9487131]
- 135.
- Seemann J, Pypaert M, Taguchi T. et al. Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science. 2002;295(5556):848–851. [PubMed: 11823640]
- 136.
- Sutterlin C, Hsu P, Mallabiabarrena A. et al. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109(3):359–369. [PubMed: 12015985]
- 137.
- Acharya U, Mallabiabarrena A, Acharya JK. et al. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell. 1998;92(2):183–192. [PubMed: 9458043]
- 138.
- Misteli T, Warren G. COP-coated vesicles are involved in the mitotic fragmentation of Golgi stacks in a cell-free system. J Cell Biol. 1994;125(2):269–282. [PMC free article: PMC2120040] [PubMed: 8163545]
- 139.
- Misteli T, Warren G. A role for tubular networks and a COP I-independent pathway in the mitotic fragmentation of Golgi stacks in a cell-free system. J Cell Biol. 1995;130(5):1027–1039. [PMC free article: PMC2120551] [PubMed: 7657690]
- 140.
- Lowe M, Rabouille C, Nakamura N. et al. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94(6):783–793. [PubMed: 9753325]
- 141.
- Kano F, Takenaka K, Yamamoto A. et al. MEK and Cdc2 kinase are sequentially required for Golgi disassembly in MDCK cells by the mitotic Xenopus extracts. J Cell Biol. 2000;149(2):357–368. [PMC free article: PMC2175170] [PubMed: 10769028]
- The Golgi Apparatus - Madame Curie Bioscience DatabaseThe Golgi Apparatus - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...