NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Transfer of mobile genetic elements between prokaryotes is limited by restriction-modification systems. Restriction-modification systems consist of a modification enzyme that epigenetically methylates a specific DNA sequence, and a restriction endonuclease (restriction enzyme) that cuts DNA lacking this epigenetic mark. These elements were discovered because they attack mobile genetic elements. However, recent studies have revealed that they are themselves mobile. In some cases, the mobility of restriction-modification systems is through symbiosis with other forms of mobile elements. In other cases, movement is unlinked to other mobile elements. The systems may insert into the genome with long and variable target duplication, or into the intergenic region of an operon. Insertion of restriction-modification systems induces other genome rearrangements such as amplification and inversion. Even a domain within a protein can be the unit of mobility: some restriction-modification system subunits that recognize a target DNA sequence contain mobile amino acid sequences that can apparently move between different domains of a protein through recombination of DNA sequences encoding them. This mobility extends the biological significance of restriction-modification systems beyond defense: the systems define, and sometimes even force, epigenetic order on a genome. The multilevel conflicts involving these mobile epigenetic elements may drive prokaryotic evolution.
Introduction: Restriction-Modification Systems in Epigenetic Conflicts
DNA methyltransferases methylate specific bases in recognition sequences and generate three types of modified base: 5-methylcytosine (m5C), N4-methylcytosine (m4C), and N6-methyladenine (m6A). This is a type of epigenetic modification because it is passed on through maintenance methylation after DNA replication.1 Epigenetic DNA methylation can be involved in gene regulation, and variation in DNA methyltransferases can potentially provide diversity in the gene expression status of the prokaryotic cell.
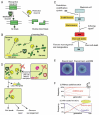
Many prokaryotic DNA methyltransferases are paired with restriction enzymes, which were first discovered through their ability to restrict phage infection.2 Restriction enzymes are DNA endonucleases that recognize specific DNA sequences and introduce a break (Fig. 1A). This activity restricts the establishment of invading DNAs that lack proper DNA methylation, such as bacteriophage DNA genomes, plasmids, and DNA fragments delivered through natural transformation machinery (Fig. 1B). The potentially lethal cleavage of cellular DNA in cells that harbor a restriction enzyme is prevented by epigenetic DNA methylation by the cognate DNA methyltransferase (Fig. 1A,B). Genes encoding the restriction enzyme and the methyltransferase are often located next to each other and form a unit called a restrictionmodification (RM) system. RM systems are classified into four types, Type I, II, III, and IV, based on their genetic and biochemical characteristics.
The primary biological significance of RM systems is often assumed to be their activity as a defense system for host cells against invading DNA such as bacteriophages. However, an RM system will not defend a bacterial cell from invasion of DNA from a bacterial cell carrying the same RM system. An RM system will attack invading DNA only when it lacks its epigenetic mark. The essence of the RM phenomena is conflict involving an epigenetic system rather than defense against invading DNA. In response to violation of epigenetic status by invasion of foreign DNA or loss of an RM system, RM systems induce DNA breakage and, consequently, reactions such as DNA damage repair, cell death and genome rearrangements (Fig. 1C,D).
RM systems are themselves mobile and can therefore be designated epigenetic mobile elements. This review introduces the mobility of RM systems first and then the conflicts between RM systems and their host cells and between RM systems themselves. These conflicts, which can involve host cell death, affect the conservation or change of the host epigenetic status.
Abundance of RM systems
RM gene homologs that have been identified in completely sequenced bacterial genomes are in the REBASE database.3 Some genomes—for example, those from Hemophilus influenzae, Methanococcus jannaschii, Helicobacter pylori, Neisseria meningitidis, Neisseria gonorrhoeae, and Xylella fastidiosa—have large numbers of RM gene homologs, although many RM gene homologs are specific to a single strain within a given species. No RM system is native to eukaryotes, although a family of viruses that use the unicellular eukaryotic alga Chlorella as a host produce RM systems.4
Types of Restriction Systems
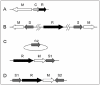
RM systems are currently classified into four types (I-IV), each with a unique mechanism for target recognition (Fig. 2).2 Type I systems consist of R, M and specificity (S) subunit genes. Formation of a multisubunit structure is necessary for modification (SM) and restriction (SMR) activities.5 Sequence recognition is determined by the target recognition domains (TRDs) in the S subunit. Each of the two TRDs, TRD1 and TRD2, recognizes one half of a bipartite target sequence.6 DNA methylation takes place within the recognition sequence, whereas the cleavage site is at a variable distance from the recognition sequence. After binding to an unmodified recognition sequence, restriction enzyme complexes are thought to translocate DNA toward themselves from both directions, in a reaction coupled to ATP hydrolysis. DNA is cleaved where two restriction enzyme complexes meet.7
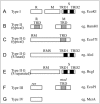
Figure 2.
Gene organization of various types of restriction-modification systems. See text for explanation. TRD: target recognition domain.
Type II systems consist of separate R and M enzymes, which independently recognize a target sequence and catalyze reactions.2,8 M proteins have amino acid sequence motifs that are common to DNA methyltransferases and are well conserved, and their target recognition domain can be easily identified.9 R proteins share much less similarity.10
RM systems of the Type IIG subgroup are defined by the presence of a single polypeptide that apparently results from a fusion of R and M proteins. This Type IIG subgroup can be further classified into two subgroups by sequence similarity to either Type II or Type I systems.11 The class similar to Type II (Fig. 2C) has the same TRD structure as typical Type II M enzymes within an RM polypeptide.2,11-13 The other class similar to Type I, such as AloI,14 has a fused S subunit in addition to the RM fusion polypeptide (Fig. 2D). As in Type I systems, the S subunit counterpart has two TRDs. Although R and M are always fused in Type IIG systems, the S subunit is sometimes separated from the RM fused gene (Fig. 2E).15
Type III systems consist of res and mod genes (Fig. 2F). The mod gene product has modification activity by itself, while the complex of the two gene products has restriction enzyme activity.16 The Mod subunit is responsible for target recognition and its TRD can be easily identified, similar to M enzymes of Type II systems.17-19
Type IV systems contain a class of enzymes that cleave DNA only when the recognition site is methylated (Fig. 2F).2 McrA, McrBC, and Mrr are prototype Escherichia coli enzymes in this class that show different restriction spectra.1,20-23
Gene Organization
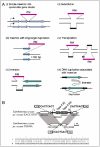
There are many variations to the simplified schemes above.3 A Type II system may have two different modification enzymes for the top and bottom strands of the recognition sequence. It may carry two different nucleases for the different strands. Some RM systems have a gene for a regulatory protein (C protein, Fig. 3A, also see below).
The relative direction of the constituent genes can vary. In the simplest case of a Type II system with one R gene and one M gene, the two genes may lie in the same orientation, or in opposite orientations.
In most cases, genes in an RM system are clustered into a single locus, but in some cases they are unlinked or in a more complex context.24 In a Type I system of Staphylococcus aureus, one pair of M and S genes lie on one genomic island, another M and S pair are on another genomic island, and an R gene is outside of both islands (Fig. 3B). The hypothesis is that the R gene product can associate with either MS pair to form a two-faced restriction enzyme that recognizes two different sequences.25 Lactococcus lactis has plasmids that carry an S gene but not an R or an M gene (Fig. 3C). The plasmid-encoded S gene products form active Type I RM systems with R and M products from genes on the chromosome.26 In Mycoplasma pulmonis, two S genes in opposing orientation flank an R gene and an M gene in the inverted orientation (Fig. 3D). Recombination between the two S genes is discussed below.27
Mobility of RM Systems Revealed from Molecular Evolutionary Analyses
Evolutionary analyses suggest that RM genes have undergone extensive horizontal transfer between different groups of microorganisms.28,29 Early studies found that close homologs occur in distantly related organisms such as Eubacteria and Archaea (archaebacteria).30 Extensive sequence alignment and phylogenetic tree construction now provide strong support for this point.31-34 Additional evidence for extensive horizontal transfer of RM genes comes from incongruencies between methyltransferase phylogenetic trees and rRNA gene trees from the same species.31 Moreover, the GC content and/or codon usage of RM genes often differs from the majority of other genes in the genome.31,35-37 This indicates that some RM genes may have joined the genome relatively recently through horizontal transfer from distantly related bacteria.
RM Systems on Mobile Genetic Elements
Genome comparisons revealed that the RM systems are mobile and involved in genome rearrangements. In some cases, mobility is acquired by carriage on other mobile elements (Table 1) such as plasmids,38-45 phages,45-52 conjugative elements/genomic islands,25,53-57 transposons,58-60 and integrons.61,62 Indeed, some RM systems are located immediately adjacent to mobility-related genes such as those for transposases, integrases, or resolvases.63-66 RM systems are also found that form a composite transposon by insertion of insertion sequence (IS) to both sides.67
Table 1.
RM systems on mobile genetic elements.
The mobile elements themselves can be lost from the genome. Carrying a Type II RM system may allow their stable maintenance through postsegregational killing, as demonstrated for plasmids.68,69 This may represent the symbiosis of two genetic elements that provides mutual benefits of maintenance and mobility.
RM Systems as Mobile Genetic Elements
In other cases, RM systems are found inserted in the genome, but not linked to a mobile element (Table 2). They sometimes insert into an operon-like gene cluster (Fig. 4A (i))70-72 by a recombination process that could be related to the DNA cleavage activity of the restriction enzyme.73 Insertion might give a competitive advantage to the operon as to the mobile elements. Some RM systems are substituted by another RM system at the same locus.70,74-76
Table 2.
Genome rearrangements associated with RM systems.
In Helicobacter pylori, insertion of several RM systems has occurred with the duplication of a long target sequence (Fig. 4A (iii)).73 The generality of this mode was confirmed by systematic comparison analysis of RM systems on the completely sequenced genomes.74 This survey also led to the discovery of RM systems with a transposon-like structure where RM genes replaced the transposase gene (Fig. 4B).74
Attack on the Host Bacterial Genome: Type II RM Systems
Works have demonstrated that RM systems also act as a watchdog maintaining epigenetic order in cells (Fig. 5). Alteration in the epigenetic status might lead to double-strand breakage of the self genome by the restriction enzyme,34,68,77,78 which can result in cell death or genome rearrangements. This may eliminate unstable cells and maintain the epigenome status.
Some Type II RM systems cause chromosomal cleavage of their host cells when their genes are eliminated, for example, by a competitor genetic element (Figs. 1D and 5A).68 When an RM system is stably maintained in a cell, the restriction enzyme does not cleave the genomic DNA because of protection through epigenetic methylation by the cognate methyltransferases. However, when the RM system is lost, the concentration of the restriction and modification enzymes is decreased through cell division,79 resulting in undermethylated sites on newly replicated chromosomes.80 The remaining restriction enzyme molecules cleave the unmethylated recognition sequence and cause cell death. The net result is survival of cells that were not invaded by the competitor. This process is called "postsegregational killing" or "genetic addiction".24 The capability of an RM system in forcing maintenance on its host can become stronger by a mutation in its methyltransferase.81
Figure 1E visualizes the effect of postsegregational killing during the formation of bacterial colony. An unstable plasmid in the bacterial cell is lost during colony formation and leads to formation of papillae (Fig. 1E, left). However, when the EcoRI RM system is present on the plasmid, no papillae are formed because the plasmid-free cells are killed (Fig. 1E, right).
Postsegregational killing occurs because of a conflict between the RM system (or the plasmid) and the host bacteria and is an example of intragenomic conflicts. A theoretical work demonstrated that starting from very few copies, a postsegregational killing gene can increase in a population in the presence of a spatial structure (Fig. 1F (ii)). In the absence of the spatial structure, the gene is quickly lost unless it is abundant at the beginning (Fig. 1F (i)).82
Post-segregational cell killing by one RM system is inhibited by the presence of another RM system recognizing the same DNA sequence, because the M protein of the latter system protects the genome from cleavage by the R protein of the former system.83 This indicates the presence of competition for recognition sequences between RM systems. Thus, a recognition sequence of RM systems defines an incompatibility group. This competition explains the individual specificity and collective diversity in RM systems' sequence recognition. The competition may be one-sided when the recognition sequence of one RM is included in the recognition sequence of the other RM.84 Another incompatibility relationship between RM systems is found in a regulatory protein that delays expression of the R protein upon entry of an RM system into a new host bacterial cell.85
Recent studies revealed a common pathway of stress-induced cell death in bacteria.86,87 Transcriptome analysis during postsegregational killing by a Type II RM system revealed its similarity to killing by several bacteriocidal antibiotics.78 Thus, RM systems switch on the death pathway intrinsic to the host bacterial cells. Gene products that program bacterial cell death, such as the restriction enzymes discussed here, are likely to work upstream of the common cell death pathway.
Attack on the Host Bacterial Genome: Type IV Restriction Systems
Several studies demonstrated that phages or plasmids carrying a DNA methyltransferase gene cannot be propagated in an strain of E. coli which harbors mcrBC, a methylated DNA specific restriction enzyme.88 Whether the block to propagation is caused by repeated methylation and subsequent cleavage of the introduced DNA,88 or to host genome methylation and its cleavage was not known. Fukuda et al. demonstrated that McrBC inhibits establishment of the gene for the DNA methyltransferase PvuII (M.PvuII, 5′CAGm4CTG) in E. coli,34 even when the gene is on a plasmid lacking its recognition sequence. This result suggests that the transferred DNA does not need to have methylated sites for McrBC-dependent inhibition,34 suggesting that host genome cleavage accompanied by cell death inhibits the establishment of the methyltransferase gene (Fig. 5B). The underlying mechanism of cell death was revealed by observing E. coli chromosomal DNA infected with lambda phage carrying the M.PvuII gene.34 Accumulation of large linear DNAs corresponding to broken chromosomes, and smaller DNAs of variable size was observed, which likely reflected chromosomal degradation. The mcrBC-dependence strongly suggests that M.PvuII-mediated chromosomal methylation triggers chromosomal cleavage by McrBC, followed by chromosomal degradation. This, in turn, indicates that inhibition of phage multiplication (restriction) is caused by host death.34 This type of conflict between DNA methyltransferase genes carried by bacteriophages and methyl-specific restriction enzymes is biologically relevant because DNA methyltransferase genes are often found in bacteriophage genomes.89-92
In addition to M.PvuII, the M.SinI (GGWm5CC) and M.MspI (m5CCGG) cause McrBC-dependent cell death, whereas M.SsoII (Cm5CNGG) does not. These results are consistent with the RmC sequence specificity of McrBC observed in vitro.93 McrBC has the potential to act as a defense system against many DNA methyltransferases with an appropriate specificity. Such conflicts between McrBC and invading epigenetic DNA methylation systems may have driven diversification of sequence recognition by the methyltransferases and by the McrBC family.94
Type II systems cause cell death when a particular mode of epigenetic DNA methylation decreases, while this Type IV system causes cell death when an epigenetic DNA methylation mode increases. Nonetheless, the result of both systems is the maintenance of an epigenetic order defined by DNA methylation (Fig. 5).
The DNA replication fork may be the site of action of McrBC, which can cleave a model DNA replication fork in vitro.77 Cleavage of a fork requires methylation on both arms and results in removal of one or both arms. Most cleavage events remove the methylated sites from the fork. This suggests that acquisition of even rare modification patterns will be recognized and rejected efficiently by modification-dependent restriction systems that recognize two sites.
Attack on the Host Bacterial Genome: Type I RM Systems
Restriction alleviation is a phenotypic decrease in restriction activity against invading DNA that can be induced by DNA-damaging agents; this also occurs constitutively in some bacterial mutants. The underlying mechanisms vary among the restriction system types.95-98 Restriction alleviation is proposed to be a mechanism for protecting chromosomes from restriction at newly generated replication forks that produce unmethylated restriction sites.5
A recent work demonstrated that a Type I restriction enzyme cleaves model replication forks at their branch point in vitro.77 Cleavage was dependent on a recognition sequence on one of the arms and was inhibited when the site was hemi-methylated. The results suggested that the enzyme binds to DNA at the recognition sequence and tracks along the DNA, cleaving when it encounters a branch point.
Fork cleavage may take place on chromosomal DNA under conditions of extra replication initiation. From an unmethylated recognition sequence, the restriction enzyme tracks on the DNA. If the fork is moving forward during replication, DNA cleavage might not occur. However, when the enzyme meets an arrested replication fork, one arm is cleaved, possibly leading to cell death or a round of repair through recombination and replication. This mechanism might lead to elimination of cells with an unstable genome and to maintenance of an intact genome.26 This hypothesized function is similar to that of programmed cell death in multicellular organisms.
Anti-Restriction Systems
The anti-restriction modification systems evolved by bacteriophages represent examples of co-evolution with the host bacterium.16 Host bacteria cells have evolved anti-restriction features, such as solitary methyltransferases that protect restriction sites from lethal attack by an RM system.81,99 Another sign of adaptation, restriction avoidance, is discussed below.
RM Systems and Genome Rearrangements
The attack of RM systems on the host genome induces repair by recombination and replication and may induce genome rearrangements (Fig. 4A). Systematic genome comparison shows the involvement of RM systems in genome rearrangements (Table 2). RM systems can be inserted into the genomes with other mobile elements (see above) or inserted by themselves with long target duplication (Fig. 4A (iii)).73,74 A large genome inversion event is seen in the neighborhood of RM insertion in Helicobacter pylori (Fig. 4A (v)).100 Activity of an RM system likely induced unequal homologous recombination at IS3 sequences in E. coli, causing genome-wide rearrangements.80 A methyltransferase gene in Helicobacter pylori is linked to an event of DNA duplication associated with inversion (Fig. 4A (vi)), a mechanism formally similar to replicative inversion in several DNA transposons.101 A Type II restriction-modification system accelerates genome and phenotype changes in bacterial experimental evolution.102
We found that the bamHI gene complex, flanked by long direct repeats, amplifies in the Bacillus subtilis chromosome in its restriction activity dependent manner.103 These results led us to propose that RM gene complexes increase in frequency in the cell population in a life cycle similar to that of a DNA virus (Fig. 1C). Insertion with long target duplication (Fig. 4A (iii), discussed above) results in formation of direct repeats flanking an RM system, giving it the potential for amplification.
Impact on Genome Evolution
The presence of an RM gene complex104 and the action of a restriction enzyme105 induce the SOS response, as does the action of a methylated DNA-specific endonuclease.106 Global mutagenesis generates heterogeneity in the cell population, similar to other stressful conditions,107 and may help the survival of the genome. These mechanisms could contribute to the evolution of restriction avoidance (discussed below) and to the inactivation of a RM gene complex. Genomes show signs of strong selection against restriction sites. Systematic avoidance of potential restriction sites (palindromes) in bacterial genomes is called restriction avoidance and has been characterized by informatics.108-110 This is likely the result of selection after host attack by RM systems. (However, evaluating the contribution, if any, of selection by restriction attacks on groups of transferred genomic DNAs is difficult.) In many genomes, restriction avoidance is more pronounced in the genome proper than in prophages.109,110 This is probably because the genome has experienced long-term selection by RM systems, while the prophages, as newcomers, have not yet undergone selection.109,110 This restriction avoidance would lead to a decrease in the virulence of the particular RM system against the genome, which is almost always beneficial for the host and can be beneficial to the RM system itself under some conditions.
DNA methylation may locally increase mutation. 5-methylcytosine shows a higher rate of deamination than cytosine. Its deamination at the C/G pair in duplex DNA results in the formation of thymine and hence the generation of a T/G mispair.111,112 Very short patch mismatch correction by a vsr gene linked to a solitary cytosine methyltransferase dcm in E. coli, can repair the mispair generated by methylation by dcm product and restore the mC/G pair.112 A homolog of the vsr gene is linked to several RM genes that produce 5-methylcytosine,3 some of which are active. Local mutagenesis induced by the action of the RM systems leads to loss of the restriction sites in the genome, which can be beneficial to the host. The anti-mutagenesis activity of Vsr homologs linked to RM systems will prevent this loss and may be of immediate benefit to the RM system.
Domain Sequence Movement in the Specificity Subunit
The above results revealed that RM systems have many features as mobile genetic elements. Recent work demonstrates that parts of RM system genes are variable and even mobile.
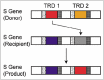
DNA sequence recognition of the Type I RM system specificity subunit is mediated by TRD1 and TRD2. These TRDs show diversity among strains, consistent with the diversification of recognition sequences.113-117 In some cases, diversification occurred by a specific genome rearrangement mechanism. In Mycoplasma pulmonis, two S genes flank the R and M genes, which are in an inverted orientation and prone to recombine with each other, resulting in TRD shuffling.27 In Lactococcus lactis, two copies of the S genes are on different plasmids, interacting through homologous recombination to create two chimeric S genes for one RM system with shuffled recognition sequences.118 S genes tolerates exchange of the sequences between TRD1 and TRD2 by circular permutation.119 Weak sequence similarity (36% identity in amino acid sequence) was detected between TRD1 and TRD2 of from different species, that is TRD1 of StySKI from Salmonella enterica and TRD2 of EcoR124I from Escherichia coli.120
Surprisingly, several sequences are shared by TRD1 and TRD2 genes at the same locus in several bacteria: these domain sequences appear to have moved between two positions within a single protein (Fig. 6).121 The gene/protein organization can be represented as x-(TRD1)-y-x-(TRD2)-y, where x and y are repeated. Movement probably occurs by recombination at these flanking DNA repeats. Lateral domain movements within a protein, which we have designated DOMO (domain movement), represent novel routes for the diversification of proteins.
Gene Regulation
RM systems possess mechanisms that tightly regulate their gene expression to suppress the potential of a lethal attack on the host bacteria. When RM systems enter a new host bacterial cell with a genome that lacks proper methylation, they avoid killing the cell by expressing the modification enzyme first (Fig. 1C).122 The restriction endonuclease and modification enzyme activities must be carefully regulated not only during RM system establishment in a new host, but also during maintenance. When RM genes are lost from a cell or the epigenetic status is disturbed, the restriction enzyme will attack the chromosome through postsegregational killing, leading to cell death or genome repair by recombination and replication. DNA fragments encoding RM systems may be released into the environment after cell death, invading other cells and establishing in the genome of a new host. This life cycle is similar to other mobile elements such as lysogenic phages and DNA transposons.
The regulatory machinery of mobile RM systems is expected to be host-independent, to bypass differences in the host factors affecting their establishment, maintenance and host attack. Thus far, regulatory mechanisms have been studied mainly at the transcriptional level,
as three main modes of regulation. One employs C-proteins,123 which specifically bind a DNA operator sequence through a helix-turn-helix (HTH) motif to temporally control expression of the restriction enzyme, the modification enzyme or both.124,125 This tight, finely tuned regulation operates via transcriptional feedback circuits.126 Moreover, C proteins can efficiently delay expression of the restriction enzyme during establishment in a new host cell.85,122 In the second type of regulation, the modification enzyme represses transcription of its own gene and occasionally stimulates restriction gene expression by DNA binding via its HTH domain.127-129 In the third mode, the coordinated expression of R-M systems depends on the methylation status of a cognate recognition site(s) in their promoter region.130,131
Recent analyses revealed regulation by intragenic reverse promoters from which antisense RNAs are transcribed.132-135
Conclusion and Perspective
RM systems, originally found to be a barrier to gene mobility, turn out to be mobile elements themselves. They are mobile epigenetic elements because they define, and sometimes even force an epigenetic status on a genome. Multilevel conflicts involving these epigenetic systems may drive prokaryotic evolution.
This model is based on laboratory experiments and genome comparisons and needs to be examined through additional experimental and theoretical studies.136 The expanding accumulation of bacterial genome sequences, especially within a species, may allow more detailed analysis. The concept of conflicts between epigenetic systems may provide information for understanding eukaryotic evolution and the origin of life.
Acknowledgments
We thank Noriko Takahashi for providing Figure 1E.
References
- 1.
- Ishikawa K, Fukuda E, Kobayashi I. Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems. DNA Res. 2010;17:325–42. doi:10.1093/dnares/dsq027. [PMC free article: PMC2993543] [PubMed: 21059708]
- 2.
- Roberts RJ, Belfort M, Bestor T, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–12. doi:10.1093/nar/gkg274. [PMC free article: PMC152790] [PubMed: 12654995]
- 3.
- Roberts RJ, Vincze T, Posfai J, et al. REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38(Database issue):D234-6. doi:10.1093/nar/gkp874. [PMC free article: PMC2808884] [PubMed: 19846593]
- 4.
- Van Etten JL, Meints RH. Giant viruses infecting algae. Annu Rev Microbiol. 1999;53:447–94. doi:10.1146/annurev.micro.53.1.447. [PubMed: 10547698]
- 5.
- Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol Mol Biol Rev. 2000;64:412–34. doi:10.1128/MMBR.64.2.412-434.2000. [PMC free article: PMC98998] [PubMed: 10839821]
- 6.
- Gough JA, Murray NE. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983;166:1–19. doi:10.1016/S0022-2836(83)80047-3. [PubMed: 6304321]
- 7.
- Studier FW, Bandyopadhyay PK. Model for how type I restriction enzymes select cleavage sites in DNA. Proc Natl Acad Sci USA. 1988;85:4677–81. doi:10.1073/pnas.85.13.4677. [PMC free article: PMC280498] [PubMed: 2838843]
- 8.
- Pingoud A, Fuxreiter M, Pingoud V, et al. Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci. 2005;62:685–707. doi:10.1007/s00018-004-4513-1. [PubMed: 15770420]
- 9.
- Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–32. doi:10.1006/jmbi.1995.0577. [PubMed: 7473738]
- 10.
- Orlowski J, Bujnicki JM. Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res. 2008;36:3552–69. doi:10.1093/nar/gkn175. [PMC free article: PMC2441816] [PubMed: 18456708]
- 11.
- Morgan RD, Dwinell EA, Bhatia TK, et al. The MmeI family: type II restriction-modification enzymes that employ single-strand modification for host protection. Nucleic Acids Res. 2009;37:5208–21. doi:10.1093/nar/gkp534. [PMC free article: PMC2731913] [PubMed: 19578066]
- 12.
- Janulaitis A, Petrusyte M, Maneliene Z, et al. Purification and properties of the Eco57I restriction endonuclease and methylase-prototypes of a new class (type IV). Nucleic Acids Res. 1992;20:6043–9. doi:10.1093/nar/20.22.6043. [PMC free article: PMC334471] [PubMed: 1334260]
- 13.
- Nakonieczna J, Kaczorowski T, Obarska-Kosinska A, et al. Functional analysis of MmeI from methanol utilizer Methylophilus methylotrophus, a subtype IIC restriction-modification enzyme related to type I enzymes. Appl Environ Microbiol. 2009;75:212–23. doi:10.1128/AEM.01322-08. [PMC free article: PMC2612229] [PubMed: 18997032]
- 14.
- Cesnaviciene E, Petrusyte M, Kazlauskiene R, et al. Characterization of AloI, a restriction-modification system of a new type. J Mol Biol. 2001;314:205–16. doi:10.1006/jmbi.2001.5049. [PubMed: 11718555]
- 15.
- Kong H. Analyzing the functional organization of a novel restriction modification system, the BcgI system. J Mol Biol. 1998;279:823–32. doi:10.1006/jmbi.1998.1821. [PubMed: 9642063]
- 16.
- Dryden DT, Murray NE, Rao DN. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–41. doi:10.1093/nar/29.18.3728. [PMC free article: PMC55918] [PubMed: 11557806]
- 17.
- Fox KL, Dowideit SJ, Erwin AL, et al. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 2007;35:5242–52. doi:10.1093/nar/gkm571. [PMC free article: PMC1976455] [PubMed: 17675301]
- 18.
- Srikhanta YN, Dowideit SJ, Edwards JL, et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog. 2009;5:e1000400. doi:10.1371/journal. ppat.1000400. [PMC free article: PMC2667262] [PubMed: 19390608]
- 19.
- Srikhanta YN, Fox KL, Jennings MP. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat Rev Microbiol. 2010;8:196–206. doi:10.1038/nrmicro2283. [PubMed: 20140025]
- 20.
- Kelleher JE, Raleigh EA. A novel activity in Escherichia coli K-12 that directs restriction of DNA modified at CG dinucleotides. J Bacteriol. 1991;173:5220–3. [PMC free article: PMC208216] [PubMed: 1830580]
- 21.
- Waite-Rees PA, Keating CJ, Moran LS, et al. Characterization and expression of the Escherichia coli Mrr restriction system. J Bacteriol. 1991;173:5207–19. [PMC free article: PMC208215] [PubMed: 1650347]
- 22.
- Mulligan EA, Dunn JJ. Cloning, purification and initial characterization of E. coli McrA, a putative 5-methylcytosine-specific nuclease. Protein Expr Purif. 2008;62:98–103. doi:10.1016/j. pep.2008.06.016. [PMC free article: PMC2900843] [PubMed: 18662788]
- 23.
- Mulligan EA, Hatchwell E, McCorkle SR, et al. Differential binding of Escherichia coli McrA protein to DNA sequences that contain the dinucleotide m5CpG. Nucleic Acids Res. 2010;38:1997–2005. doi:10.1093/nar/gkp1120. [PMC free article: PMC2847215] [PubMed: 20015968]
- 24.
- Kobayashi I. Restriction-modification systems as minimal forms of life. Restriction endonucleases. 2004:19–62.
- 25.
- Kuroda M, Ohta T, Uchiyama I, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–40. doi:10.1016/S0140-6736(00)04403-2. [PubMed: 11418146]
- 26.
- Schouler C, Gautier M, Ehrlich SD, et al. Combinational variation of restriction modification specificities in Lactococcus lactis. Mol Microbiol. 1998;28:169–78. doi:10.1046/ j.1365-2958.1998.00787.x. [PubMed: 9593305]
- 27.
- Dybvig K, Sitaraman R, French CT. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc Natl Acad Sci USA. 1998;95:13923–8. doi:10.1073/pnas.95.23.13923. [PMC free article: PMC24968] [PubMed: 9811902]
- 28.
- Kobayashi I, Nobusato A, Kobayashi-Takahashi N, et al. Shaping the genome-restriction-modification systems as mobile genetic elements. Curr Opin Genet Dev. 1999;9:649–56. doi:10.1016/S0959-437X(99)00026-X. [PubMed: 10607611]
- 29.
- Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–56. doi:10.1093/nar/29.18.3742. [PMC free article: PMC55917] [PubMed: 11557807]
- 30.
- Nölling J, de Vos WM. Characterization of the archaeal, plasmid-encoded type II restriction-modification system MthTI from Methanobacterium thermoformicicum THF: homology to the bacterial NgoPII system from Neisseria gonorrhoeae. J Bacteriol. 1992;174:5719–26. [PMC free article: PMC206520] [PubMed: 1512204]
- 31.
- Nobusato A, Uchiyama I, Kobayashi I. Diversity of restriction-modification gene homologues in Helicobacter pylori. Gene. 2000;259:89–98. doi:10.1016/S0378-1119(00)00455-8. [PubMed: 11163966]
- 32.
- Jeltsch A, Kroger M, Pingoud A. Evidence for an evolutionary relationship among type-II restriction endonucleases. Gene. 1995;160:7–16. doi:10.1016/0378-1119(95)00181-5. [PubMed: 7628720]
- 33.
- Bujnicki JM, Radlinska M. Molecular phylogenetics of DNA 5mC-methyltransferases. Acta Microbiol Pol. 1999;48:19–30. [PubMed: 10467693]
- 34.
- Fukuda E, Kaminska KH, Bujnicki JM, et al. Cell death upon epigenetic genome methylation: a novel function of methyl-specific deoxyribonucleases. Genome Biol. 2008;9:R163. doi:10.1186/gb-2008-9-11-r163. [PMC free article: PMC2614495] [PubMed: 19025584]
- 35.
- Jeltsch A, Pingoud A. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J Mol Evol. 1996;42:91–6. doi:10.1007/BF02198833. [PubMed: 8919860]
- 36.
- Mrázek J, Karlin S. Detecting alien genes in bacterial genomes. Ann N Y Acad Sci. 1999;870:314–29. doi:10.1111/j.1749-6632.1999.tb08893.x. [PubMed: 10415493]
- 37.
- Parkhill J, Achtman M, James KD, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–6. doi:10.1038/35006655. [PubMed: 10761919]
- 38.
- Karyagina AS, Lunin VG, Degtyarenko KN, et al. Analysis of the nucleotide and derived amino acid sequences of the SsoII restriction endonuclease and methyltransferase. Gene. 1993;124:13–9. doi:10.1016/0378-1119(93)90756-S. [PubMed: 7916706]
- 39.
- Lubys A, Janulaitis A. Cloning and analysis of the plasmid-borne genes encoding the Bsp6I restriction and modification enzymes. Gene. 1995;157:25–9. doi:10.1016/0378-1119(94)00795-T. [PubMed: 7607501]
- 40.
- Glatman LI, Moroz AF, Yablokova MB, et al. A novel plasmid-mediated DNA restriction-modification system in E. coli. Plasmid. 1980;4:350–1. doi:10.1016/0147-619X(80)90072-4. [PubMed: 6261280]
- 41.
- Hinkle NF, Miller RV. pMG7-mediated restriction of Pseudomonas aeruginosa phage DNAs is determined by a class II restriction endonuclease. Plasmid. 1979;2:387–93. doi:10.1016/0147-619X (79)90022-2. [PubMed: 113797]
- 42.
- Greene PJ, Gupta M, Boyer HW, et al. Sequence analysis of the DNA encoding the Eco RI endonuclease and methylase. J Biol Chem. 1981;256:2143–53. [PubMed: 6257703]
- 43.
- Kosykh VG, Buryanov YI, Bayev AA. Molecular cloning of EcoRII endonuclease and methylase genes. Mol Gen Genet. 1980;178:717–8. doi:10.1007/BF00337884. [PubMed: 6248737]
- 44.
- Calvin Koons MD, Blumenthal RM. Characterization of pPvu1, the autonomous plasmid from Proteus vulgaris that carries the genes of the PvuII restriction-modification system. Gene. 1995;157:73–9. doi:10.1016/0378-1119(94)00618-3. [PubMed: 7607530]
- 45.
- Hümbelin M, Suri B, Rao DN, et al. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J Mol Biol. 1988;200:23–9. doi:10.1016/0022-2836(88)90330-0. [PubMed: 2837577]
- 46.
- Borriss M, Lombardot T, Glockner FO, et al. Genome and proteome characterization of the psychrophilic Flavobacterium bacteriophage 11b. Extremophiles. 2007;11:95–104. doi:10.1007/ s00792-006-0014-5. [PubMed: 16932843]
- 47.
- Miner Z, Hattman S. Molecular cloning, sequencing, and mapping of the bacteriophage T2 dam gene. J Bacteriol. 1988;170:5177–84. [PMC free article: PMC211587] [PubMed: 3053648]
- 48.
- Dempsey RM, Carroll D, Kong H, et al. Sau42I, a BcgI-like restriction-modification system encoded by the Staphylococcus aureus quadruple-converting phage Phi42. Microbiology. 2005;151:1301–11. doi:10.1099/mic.0.27646-0. [PubMed: 15817797]
- 49.
- Ohshima H, Matsuoka S, Asai K, et al. Molecular organization of intrinsic restriction and modification genes BsuM of Bacillus subtilis Marburg. J Bacteriol. 2002;184:381–9. doi:10.1128/ JB.184.2.381-389.2002. [PMC free article: PMC139560] [PubMed: 11751814]
- 50.
- Hendrix RW, Smith MC, Burns RN, et al. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–7. doi:10.1073/pnas.96.5.2192. [PMC free article: PMC26759] [PubMed: 10051617]
- 51.
- Kita K, Kawakami H, Tanaka H. Evidence for horizontal transfer of the EcoT38I restriction-modification gene to chromosomal DNA by the P2 phage and diversity of defective P2 prophages in Escherichia coli TH38 strains. J Bacteriol. 2003;185:2296–305. doi:10.1128/JB.185.7.2296-2305.2003. [PMC free article: PMC151499] [PubMed: 12644501]
- 52.
- Kita K, Tsuda J, Kato T, et al. Evidence of horizontal transfer of the EcoO109I restriction-modification gene to Escherichia coli chromosomal DNA. J Bacteriol. 1999;181:6822–7. [PMC free article: PMC94149] [PubMed: 10542186]
- 53.
- Euler CW, Ryan PA, Martin JM, et al. M. SpyI, a DNA methyltransferase encoded on a mefA chimeric element, modifies the genome of Streptococcus pyogenes. J Bacteriol. 2007;189:1044–54. doi:10.1128/JB.01411-06. [PMC free article: PMC1797290] [PubMed: 17085578]
- 54.
- Sampath J, Vijayakumar MN. Identification of a DNA cytosine methyltransferase gene in conjugative transposon Tn5252. Plasmid. 1998;39:63–76. doi:10.1006/plas.1997.1316. [PubMed: 9473447]
- 55.
- O'Sullivan DJ, Zagula K, Klaenhammer TR. In vivo restriction by LlaI is encoded by three genes, arranged in an operon with llaIM, on the conjugative Lactococcus plasmid pTR2030. J Bacteriol. 1995;177:134–43. [PMC free article: PMC176565] [PubMed: 7528201]
- 56.
- Burrus V, Bontemps C, Decaris B, et al. Characterization of a novel type II restriction-modification system, Sth368I, encoded by the integrative element ICESt1 of Streptococcus thermophilus CNRZ368. Appl Environ Microbiol. 2001;67:1522–8. doi:10.1128/AEM.67.4.1522-1528.2001. [PMC free article: PMC92764] [PubMed: 11282600]
- 57.
- Böltner D, MacMahon C, Pembroke JT, et al. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J Bacteriol. 2002;184:5158–69. doi:10.1128/ JB.184.18.5158-5169.2002. [PMC free article: PMC135318] [PubMed: 12193633]
- 58.
- van Zyl LJ, Deane SM, Louw LA, et al. Presence of a family of plasmids (29 to 65 kilobases) with a 26-kilobase common region in different strains of the sulfur-oxidizing bacterium Acidithiobacillus caldus. Appl Environ Microbiol. 2008;74:4300–8. doi:10.1128/AEM.00864-08. [PMC free article: PMC2493190] [PubMed: 18515486]
- 59.
- Stiens M, Becker A, Bekel T, et al. Comparative genomic hybridisation and ultrafast pyrosequencing revealed remarkable differences between the Sinorhizobium meliloti genomes of the model strain Rm1021 and the field isolate SM11. J Biotechnol. 2008;136:31–7. doi:10.1016/j. jbiotec.2008.04.014. [PubMed: 18562031]
- 60.
- Rochepeau P, Selinger LB, Hynes MF. Transposon-like structure of a new plasmid-encoded restriction- modification system in Rhizobium leguminosarum VF39SM. Mol Gen Genet. 1997;256:387–96. doi:10.1007/s004380050582. [PubMed: 9393436]
- 61.
- Heidelberg JF, Eisen JA, Nelson WC, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–83. doi:10.1038/35020000. [PMC free article: PMC8288016] [PubMed: 10952301]
- 62.
- Rowe-Magnus DA, Guerout AM, Ploncard P, et al. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc Natl Acad Sci USA. 2001;98:652–7. doi:10.1073/pnas.98.2.652. [PMC free article: PMC14643] [PubMed: 11209061]
- 63.
- Antonenko V, Pawlow V, Heesemann J, et al. Characterization of a novel unique restriction-modification system from Yersinia enterocolitica O:8 1B. FEMS Microbiol Lett. 2003;219:249–52. doi:10.1016/S0378-1097(03)00047-8. [PubMed: 12620628]
- 64.
- Brassard S, Paquet H, Roy PH. A transposon-like sequence adjacent to the AccI restriction-modification operon. Gene. 1995;157:69–72. doi:10.1016/0378-1119(94)00734-A. [PubMed: 7607529]
- 65.
- Naderer M, Brust JR, Knowle D, et al. Mobility of a restriction-modification system revealed by its genetic contexts in three hosts. J Bacteriol. 2002;184:2411–9. doi:10.1128/ JB.184.9.2411-2419.2002. [PMC free article: PMC135005] [PubMed: 11948154]
- 66.
- Anton BP, Heiter DF, Benner JS, et al. Cloning and characterization of the Bg/II restriction-modification system reveals a possible evolutionary footprint. Gene. 1997;187:19–27. doi:10.1016/ S0378-1119(96)00638-5. [PubMed: 9073062]
- 67.
- Takahashi N, Ohashi S, Sadykov MR, et al. IS-Linked Movement of a Restriction-Modification System. PLoS ONE. 2011;6:e16554. doi:10.1371/journal.pone.0016554. [PMC free article: PMC3031569] [PubMed: 21305031]
- 68.
- Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–9. doi:10.1126/science.7846533. [PubMed: 7846533]
- 69.
- Naito Y, Naito T, Kobayashi I. Selfish restriction modification genes: resistance of a resident R/M plasmid to displacement by an incompatible plasmid mediated by host killing. Biol Chem. 1998;379:429–36. doi:10.1515/bchm.1998.379.4-5.429. [PubMed: 9628334]
- 70.
- Sekizaki T, Otani Y, Osaki M, et al. Evidence for horizontal transfer of SsuDAT1I restriction-modification genes to the Streptococcus suis genome. J Bacteriol. 2001;183:500–11. doi:10.1128/ JB.183.2.500-511.2001. [PMC free article: PMC94905] [PubMed: 11133943]
- 71.
- Claus H, Friedrich A, Frosch M, et al. Differential distribution of novel restriction-modification systems in clonal lineages of Neisseria meningitidis. J Bacteriol. 2000;182:1296–303. doi:10.1128/ JB.182.5.1296-1303.2000. [PMC free article: PMC94415] [PubMed: 10671450]
- 72.
- Stein DC, Gunn JS, Piekarowicz A. Sequence similarities between the genes encoding the S. NgoI and HaeII restriction/modification systems. Biol Chem. 1998;379:575–8. [PubMed: 9628358]
- 73.
- Nobusato A, Uchiyama I, Ohashi S, et al. Insertion with long target duplication: a mechanism for gene mobility suggested from comparison of two related bacterial genomes. Gene. 2000;259:99–108. doi:10.1016/S0378-1119(00)00456-X. [PubMed: 11163967]
- 74.
- Furuta Y, Abe K, Kobayashi I. Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res. 2010;38:2428–43. doi:10.1093/nar/gkp1226. [PMC free article: PMC2853133] [PubMed: 20071371]
- 75.
- Sibley MH, Raleigh EA. Cassette-like variation of restriction enzyme genes in Escherichia coli C and relatives. Nucleic Acids Res. 2004;32:522–34. doi:10.1093/nar/gkh194. [PMC free article: PMC373321] [PubMed: 14744977]
- 76.
- Miller WG, Pearson BM, Wells JM, et al. Diversity within the Campylobacter jejuni type I restriction-modification loci. Microbiology. 2005;151:337–51. doi:10.1099/ mic.0.27327-0. [PubMed: 15699185]
- 77.
- Ishikawa K, Handa N, Kobayashi I. Cleavage of a model DNA replication fork by a Type I restriction endonuclease. Nucleic Acids Res. 2009;37:3531–44. doi:10.1093/nar/gkp214. [PMC free article: PMC2699502] [PubMed: 19357093]
- 78.
- Asakura Y, Kobayashi I. From damaged genome to cell surface: transcriptome changes during bacterial cell death triggered by loss of a restriction-modification gene complex. Nucleic Acids Res. 2009;37:3021–31. doi:10.1093/nar/gkp148. [PMC free article: PMC2685091] [PubMed: 19304752]
- 79.
- Ichige A, Kobayashi I. Stability of EcoRI restriction-modification enzymes in vivo differentiates the EcoRI restriction-modification system from other postsegregational cell killing systems. J Bacteriol. 2005;187:6612–21. doi:10.1128/JB.187.19.6612-6621.2005. [PMC free article: PMC1251573] [PubMed: 16166522]
- 80.
- Handa N, Nakayama Y, Sadykov M, et al. Experimental genome evolution: large-scale genome rearrangements associated with resistance to replacement of a chromosomal restriction-modification gene complex. Mol Microbiol. 2001;40:932–40. doi:10.1046/j.1365-2958.2001.02436.x. [PubMed: 11401700]
- 81.
- Ohno S, Handa N, Watanabe-Matsui M, et al. Maintenance forced by a restriction-modification system can be modulated by a region in its modification enzyme not essential for methyltransferase activity. J Bacteriol. 2008;190:2039–49. doi:10.1128/JB.01319-07. [PMC free article: PMC2258900] [PubMed: 18192396]
- 82.
- Mochizuki A, Yahara K, Kobayashi I, et al. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics. 2006;172:1309–23. doi:10.1534/genetics.105.042895. [PMC free article: PMC1456228] [PubMed: 16299387]
- 83.
- Kusano K, Naito T, Handa N, et al. Restriction-modification systems as genomic parasites in competition for specific sequences. Proc Natl Acad Sci USA. 1995;92:11095–9. doi:10.1073/ pnas.92.24.11095. [PMC free article: PMC40578] [PubMed: 7479944]
- 84.
- Chinen A, Naito Y, Handa N, et al. Evolution of sequence recognition by restriction-modification enzymes: selective pressure for specificity decrease. Mol Biol Evol. 2000;17:1610–9. [PubMed: 11070049]
- 85.
- Nakayama Y, Kobayashi I. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc Natl Acad Sci USA. 1998;95:6442–7. doi:10.1073/pnas.95.11.6442. [PMC free article: PMC27783] [PubMed: 9600985]
- 86.
- Kohanski MA, Dwyer DJ, Hayete B, et al. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi:10.1016/j.cell.2007.06.049. [PubMed: 17803904]
- 87.
- Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–35. doi:10.1038/nrmicro2333. [PMC free article: PMC2896384] [PubMed: 20440275]
- 88.
- Blumenthal RM, Cotterman MM. Isolation of mutants in a DNA methyltransferase through mcrB-mediated restriction. Gene. 1988;74:271–3. doi:10.1016/0378-1119(88)90301-0. [PubMed: 2854810]
- 89.
- Iida S, Meyer J, Bachi B, et al. DNA restriction-modification genes of phage P1 and plasmid p15B. Structure and in vitro transcription. J Mol Biol. 1983;165:1–18. doi:10.1016/ S0022-2836(83)80239-3. [PubMed: 6302279]
- 90.
- Noyer-Weidner M, Walter J, Terschuren PA, et al. M. phi 3TII: a new monospecific DNA (cytosine-C5) methyltransferase with pronounced amino acid sequence similarity to a family of adenine-N6-DNA-methyltransferases. Nucleic Acids Res. 1994;22:5517–23. doi:10.1093/ nar/22.24.5517. [PMC free article: PMC332121] [PubMed: 7816649]
- 91.
- Lange C, Noyer-Weidner M, Trautner TA, et al. M. H2I, a multispecific 5C-DNA methyltransferase encoded by Bacillus amyloliquefaciens phage H2. Gene. 1991;100:213–8. doi:10.1016/ 0378-1119(91)90369-M. [PubMed: 2055471]
- 92.
- Behrens B, Noyer-Weidner M, Pawlek B, et al. Organization of multispecific DNA methyltransferases encoded by temperate Bacillus subtilis phages. EMBO J. 1987;6:1137–42. [PMC free article: PMC553513] [PubMed: 3109889]
- 93.
- Sutherland E, Coe L, Raleigh EA. McrBC: a multisubunit GTP-dependent restriction endonuclease. J Mol Biol. 1992;225:327–48. doi:10.1016/0022-2836(92)90925-A. [PubMed: 1317461]
- 94.
- Warren RA. Modified bases in bacteriophage DNAs. Annu Rev Microbiol. 1980;34:137–58. doi:10.1146/annurev.mi.34.100180.001033. [PubMed: 7002022]
- 95.
- Doronina VA, Murray NE. The proteolytic control of restriction activity in Escherichia coli K-12. Mol Microbiol. 2001;39:416–28. doi:10.1046/j.1365-2958.2001.02232.x. [PubMed: 11136462]
- 96.
- Makovets S, Doronina VA, Murray NE. Regulation of endonuclease activity by proteolysis prevents breakage of unmodified bacterial chromosomes by type I restriction enzymes. Proc Natl Acad Sci USA. 1999;96:9757–62. doi:10.1073/pnas.96.17.9757. [PMC free article: PMC22283] [PubMed: 10449767]
- 97.
- Makovets S, Powell LM, Titheradge AJ, et al. Is modification sufficient to protect a bacterial chromosome from a resident restriction endonuclease? Mol Microbiol. 2004;51:135–47. doi:10.1046/ j.1365-2958.2003.03801.x. [PubMed: 14651617]
- 98.
- Seidel R, Bloom JG, van Noort J, et al. Dynamics of initiation, termination and reinitiation of DNA translocation by the motor protein EcoR124I. EMBO J. 2005;24:4188–97. doi:10.1038/sj.emboj.7600881. [PMC free article: PMC1356320] [PubMed: 16292342]
- 99.
- Takahashi N, Naito Y, Handa N, et al. A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction-modification gene complex. J Bacteriol. 2002;184:6100–8. doi:10.1128/JB.184.22.6100-6108.2002. [PMC free article: PMC151934] [PubMed: 12399478]
- 100.
- Alm RA, Ling LS, Moir DT, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–80. doi:10.1038/16495 . [PubMed: 9923682]
- 101.
- Furuta Y, Kawai M, Yahara K, et al. Birth and death of genes linked to chromosomal inversion. Proc Natl Acad Sci USA. 2011;108:1501–6. doi:10.1073/pnas.1012579108. [PMC free article: PMC3029772] [PubMed: 21212362]
- 102.
- Asakura Y, Kojima H, Kobayashi I. Evolutionary genome engineering using a restriction-modification system. Nucleic Acids Res. 2011 Aug. 12 [Epub ahead of print] [PMC free article: PMC3203608] [PubMed: 21785135]
- 103.
- Sadykov M, Asami Y, Niki H, et al. Multiplication of a restriction-modification gene complex. Mol Microbiol. 2003;48:417–27. doi:10.1046/j.1365-2958.2003.03464.x. [PubMed: 12675801]
- 104.
- Handa N, Ichige A, Kusano K, et al. Cellular responses to postsegregational killing by restriction-modification genes. J Bacteriol. 2000;182:2218–29. doi:10.1128/JB.182.8.2218-2229.2000. [PMC free article: PMC111271] [PubMed: 10735865]
- 105.
- Heitman J, Model P. SOS induction as an in vivo assay of enzyme-DNA interactions. Gene. 1991;103:1–9. doi:10.1016/0378-1119(91)90383-M. [PubMed: 1908806]
- 106.
- Heitman J, Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J Bacteriol. 1987;169:3243–50. [PMC free article: PMC212376] [PubMed: 3036779]
- 107.
- Higgins NP. Death and transfiguration among bacteria. Trends Biochem Sci. 1992;17:207–11. doi:10.1016/0968-0004(92)90376-K. [PubMed: 1323887]
- 108.
- Gelfand MS, Koonin EV. Avoidance of palindromic words in bacterial and archaeal genomes: a close connection with restriction enzymes. Nucleic Acids Res. 1997;25:2430–9. doi:10.1093/ nar/25.12.2430. [PMC free article: PMC1995031] [PubMed: 9171096]
- 109.
- Rocha EP, Danchin A, Viari A. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 2001;11:946–58. doi:10.1101/ gr.GR-1531RR. [PubMed: 11381024]
- 110.
- Rocha EP, Viari A, Danchin A. Oligonucleotide bias in Bacillus subtilis: general trends and taxonomic comparisons. Nucleic Acids Res. 1998;26:2971–80. doi:10.1093/nar/26.12.2971. [PMC free article: PMC147636] [PubMed: 9611243]
- 111.
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis: Amer Society for Microbiology. 1995
- 112.
- Lieb M. Spontaneous mutation at a 5-methylcytosine hotspot is prevented by very short patch (VSP) mismatch repair. Genetics. 1991;128:23–7. [PMC free article: PMC1204449] [PubMed: 1829427]
- 113.
- Fuller-Pace FV, Bullas LR, Delius H, et al. Genetic recombination can generate altered restriction specificity. Proc Natl Acad Sci USA. 1984;81:6095–9. doi:10.1073/pnas.81.19.6095. [PMC free article: PMC391866] [PubMed: 6091134]
- 114.
- Gubler M, Braguglia D, Meyer J, et al. Recombination of constant and variable modules alters DNA sequence recognition by type IC restriction-modification enzymes. EMBO J. 1992;11:233–40. [PMC free article: PMC556444] [PubMed: 1740108]
- 115.
- Gann AA, Campbell AJ, Collins JF, et al. Reassortment of DNA recognition domains and the evolution of new specificities. Mol Microbiol. 1987;1:13–22. doi:10.1111/j.1365-2958.1987. tb00521.x. [PubMed: 2838725]
- 116.
- Tsuru T, Kawai M, Mizutani-Ui Y, et al. Evolution of paralogous genes: Reconstruction of genome rearrangements through comparison of multiple genomes within Staphylococcus aureus. Mol Biol Evol . 2006;23:1269–85. doi:10.1093/molbev/msk013. [PubMed: 16601000]
- 117.
- Waldron DE, Lindsay JA. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol. 2006;188:5578–85. doi:10.1128/JB.00418-06. [PMC free article: PMC1540015] [PubMed: 16855248]
- 118.
- O'Sullivan D, Twomey DP, Coffey A, et al. Novel type I restriction specificities through domain shuffling of HsdS subunits in Lactococcus lactis. Mol Microbiol. 2000;36:866–75. doi:10.1046/j.1365-2958.2000.01901.x. [PubMed: 10844674]
- 119.
- Janscak P, Bickle TA. The DNA recognition subunit of the type IB restriction-modification enzyme EcoAI tolerates circular permutions of its polypeptide chain. J Mol Biol. 1998;284:937–48. doi:10.1006/jmbi.1998.2250. [PubMed: 9837717]
- 120.
- Thorpe PH, Ternent D, Murray NE. The specificity of sty SKI, a type I restriction enzyme, implies a structure with rotational symmetry. Nucleic Acids Res. 1997;25:1694–700. doi:10.1093/ nar/25.9.1694. [PMC free article: PMC146652] [PubMed: 9108149]
- 121.
- Furuta Y, Kawai M, Uchiyama I, et al. Domain movement within a gene: A novel evolutionary mechanism for protein diversification. PLoS ONE. 2011;6:e18819. doi:10.1371/ journal.pone.0018819. [PMC free article: PMC3077401] [PubMed: 21533192]
- 122.
- Mruk I, Blumenthal RM. Real-time kinetics of restriction-modification gene expression after entry into a new host cell. Nucleic Acids Res. 2008;36:2581–93. doi:10.1093/nar/gkn097. [PMC free article: PMC2377437] [PubMed: 18334533]
- 123.
- Tao T, Bourne JC, Blumenthal RM. A family of regulatory genes associated with type II restriction-modification systems. J Bacteriol. 1991;173:1367–75. [PMC free article: PMC207272] [PubMed: 1995588]
- 124.
- Knowle D, Lintner RE, Touma YM, et al. Nature of the promoter activated by C. PvuII, an unusual regulatory protein conserved among restriction-modification systems. J Bacteriol. 2005;187:488–97. doi:10.1128/JB.187.2.488-497.2005. [PMC free article: PMC543531] [PubMed: 15629920]
- 125.
- Vijesurier RM, Carlock L, Blumenthal RM, et al. Role and mechanism of action of C. PvuII, a regulatory protein conserved among restriction-modification systems. J Bacteriol. 2000;182:477–87. doi:10.1128/JB.182.2.477-487.2000. [PMC free article: PMC94299] [PubMed: 10629196]
- 126.
- Mruk I, Rajesh P, Blumenthal RM. Regulatory circuit based on autogenous activation-repression: roles of C-boxes and spacer sequences in control of the PvuII restriction-modification system. Nucleic Acids Res. 2007;35:6935–52. doi:10.1093/nar/gkm837. [PMC free article: PMC2175313] [PubMed: 17933763]
- 127.
- Som S, Friedman S. Regulation of EcoRII methyltransferase: effect of mutations on gene expression and in vitro binding to the promoter region. Nucleic Acids Res. 1994;22:5347–53. doi:10.1093/nar/22.24.5347. [PMC free article: PMC332081] [PubMed: 7816624]
- 128.
- Karyagina A, Shilov I, Tashlitskii V, et al. Specific binding of sso II DNA methyltransferase to its promoter region provides the regulation of sso II restriction-modification gene expression. Nucleic Acids Res. 1997;25:2114–20. doi:10.1093/nar/25.11.2114. [PMC free article: PMC146720] [PubMed: 9153310]
- 129.
- Fedotova EA, Protsenko AS, Zakharova MV, et al. SsoII-like DNA-methyltransferase Ecl18kI: interaction between regulatory and methylating functions. Biochemistry (Mosc). 2009;74:85–91. doi:10.1134/S0006297909010131. [PubMed: 19232054]
- 130.
- Beletskaya IV, Zakharova MV, Shlyapnikov MG, et al. DNA methylation at the CfrBI site is involved in expression control in the CfrBI restriction-modification system. Nucleic Acids Res. 2000;28:3817–22. doi:10.1093/nar/28.19.3817. [PMC free article: PMC110769] [PubMed: 11000275]
- 131.
- Christensen LL, Josephsen J. The methyltransferase from the LlaDII restriction-modification system influences the level of expression of its own gene. J Bacteriol. 2004;186:287–95. doi:10.1128/JB.186.2.287-295.2004. [PMC free article: PMC305755] [PubMed: 14702296]
- 132.
- Liu Y, Ichige A, Kobayashi I. Regulation of the EcoRI restriction-modification system: Identification of ecoRIM gene promoters and their upstream negative regulators in the ecoRIR gene. Gene. 2007;400:140–9. doi:10.1016/j.gene.2007.06.006. [PubMed: 17618069]
- 133.
- Mruk I, Liu Y, Ge L, et al. Antisense RNA associated with biological regulation of a restriction-modification system. Nucleic Acids Res. 2011 [PMC free article: PMC3141266] [PubMed: 21459843]
- 134.
- Nagornykh M, Zakharova M, Protsenko A, et al. Regulation of gene expression in restriction-modification system Eco29kI. Nucleic Acids Res. 2011 [PMC free article: PMC3113576] [PubMed: 21310712]
- 135.
- Liu Y, Kobayashi I. Negative regulation of the EcoRI restriction enzyme gene is associated with intragenic reverse promoters. J Bacteriol. 2007;189:6928–35. doi:10.1128/JB.00127-07. [PMC free article: PMC2045195] [PubMed: 17616602]
- 136.
- Yahara K, Fukuyo M, Sasaki A, et al. Evolutionary maintenance of selfish homing endonuclease genes in the absence of horizontal transfer. Proc Natl Acad Sci USA. 2009;106:18861–6. doi:10.1073/pnas.0908404106. [PMC free article: PMC2773979] [PubMed: 19837694]
- 137.
- Chinen A, Uchiyama I, Kobayashi I. Comparison between Pyrococcus horikoshii and Pyrococcus abyssi genome sequences reveals linkage of restriction-modification genes with large genome polymorphisms. Gene. 2000;259:109–21. doi:10.1016/S0378-1119(00)00459-5. [PubMed: 11163968]
- Introduction: Restriction-Modification Systems in Epigenetic Conflicts
- Abundance of RM systems
- Types of Restriction Systems
- Gene Organization
- Mobility of RM Systems Revealed from Molecular Evolutionary Analyses
- RM Systems on Mobile Genetic Elements
- RM Systems as Mobile Genetic Elements
- Attack on the Host Bacterial Genome: Type II RM Systems
- Attack on the Host Bacterial Genome: Type IV Restriction Systems
- Attack on the Host Bacterial Genome: Type I RM Systems
- Anti-Restriction Systems
- RM Systems and Genome Rearrangements
- Impact on Genome Evolution
- Domain Sequence Movement in the Specificity Subunit
- Gene Regulation
- Conclusion and Perspective
- Acknowledgments
- References
- Restriction-Modification Systems as Mobile Epigenetic Elements - Madame Curie Bi...Restriction-Modification Systems as Mobile Epigenetic Elements - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...