RefSeq non-redundant proteins
Related documentation
Background and Scope
A new type of RefSeq protein record which represents non-redundant protein sequences was introduced in mid-2013. This record type was introduced to address a growing issue with redundancy in the Prokaryotic RefSeq protein dataset that coincided with a significant increase in bacterial genome submissions from individual isolates and closely related bacterial strains. For example, a large number of high-quality bacterial genomes may be submitted during a disease outbreak. The submitted sequences may reflect pathogen evolution during the course of the outbreak but the majority of the encoded proteins from these genomes may be identical to each other. As RefSeq includes these genomes, per community requests, this resulted in increased redundancy. By representing identical proteins using a single non-redundant protein accession number (with the prefix 'WP_'), redundancy in the database is significantly reduced.
Non-redundant RefSeq protein records are currently provided for archaeal and bacterial RefSeq genomes, with the exception of selected reference genomes, by the NCBI prokaryotic genome annotation pipeline. This scope definition may change in the future to include additional RefSeq sub-kingdoms or other organism groups and some GenBank conceptual translation protein records may provide cross-links to RefSeq non-redundant proteins.
When the NCBI genome annotation pipeline annotates a bacterial protein that is 100% identical and the same length as an existing non-redundant protein, NCBI will annotate that protein on the genome by referencing the WP_ accession in the annotated CDS feature. Any annotation of protein function on the genome record, such as the protein name, will be inherited from the independently-maintained non-redundant protein record. Non-redundant protein records always represent one exact sequence that has been observed once or many times in different strains or species. These records will always have a version number of '1' because the sequence will never be changed, although the name and other descriptive annotation will be maintained and updated. Individual non-redundant protein records will be suppressed if that identical protein is no longer found on any RefSeq genome.
Because a non-redundant protein sequence may be found in RefSeq genomes from multiple species, the organism information provided on the protein record reflects the lowest-common taxonomic node ranging from the genus species level to super-kingdom. A non-redundant protein record that provides organism information at the level of a genus, family, or even super-kingdom does not mean that the protein is found in all RefSeq genomes below that taxonomic classification. It only indicates that the protein is found in more than one genome of different species for which the genus, family, or super-kingdom classification is the lowest common taxonomic node. Generally, identical proteins found in several species are highly conserved or the result of lateral gene transfer (either by recombination or plasmids and phages), but in some cases, this may be an indicator that the organism was misclassified in the submitted genomic sequence data which is the source of the RefSeq genome. An additional advantage of this approach to genome annotation is that it reveals these kinds of problems in the input sequences, helping to guide ongoing RefSeq curation as we correct these issues over time.
Reference Genomes and Proteomes
When an important representative genome exists, such as Escherichia coli K12 substr. MG1655, the reference genome will be annotated with strain-specific reference protein accessions with the NP_ or YP_ prefix. These protein records will refer to the matching non-redundant protein record (WP_) in the sequence block. Thus, reference proteome records (NP_ or YP_) will continue to define taxonomically-oriented sets of proteins and will track sequence changes over time. If a reference protein record is revised by curation and its version number changes, it will now refer to a different non-redundant (WP_) record that is identical to the updated reference protein sequence.
Record description in GenPept format
- Accession format: WP_ + 9 digits; the underscore '_' is part of the accession number.
- Version number: the version number is always 1.
- DEFINITION line: the definition line includes the protein name followed by an organism name in square brackets ([ ]). The organism may be identified at the level of species to super-kingdom and does not reflect strain-specific information. This line may include a prefix 'MULTISPECIES:' if the protein record is annotated on genomes from different species. If the protein is annotated on genomes from different super-kingdoms, then each super-kingdom name is provided in separate square brackets. Please see the examplesbelow.
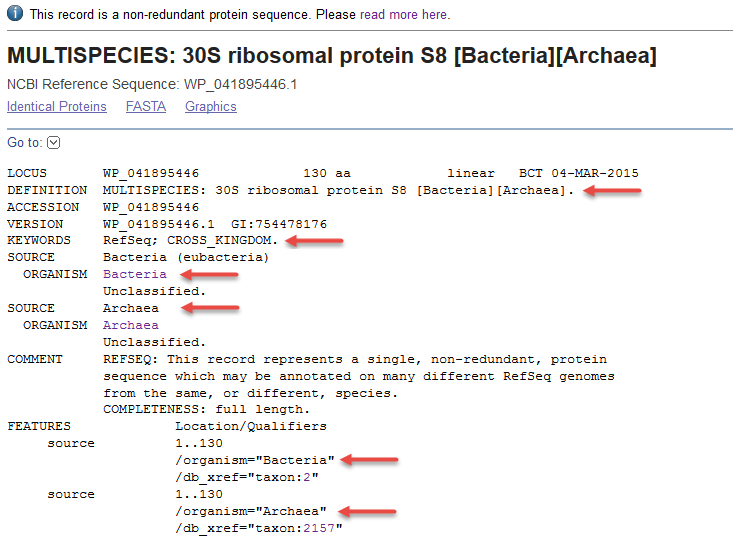
- KEYWORDS: RefSeq; CROSS_KINGDOM (optional, example WP_041895446.1)
- DBSOURCE: this line is not provided.
- ORGANISM: this line reports information at the level of super-kingdom to species and does not include strain-specific information.
- The genus species name is represented when the protein is found on one to multiple strains or isolates of the same species (example WP_000091939.1).
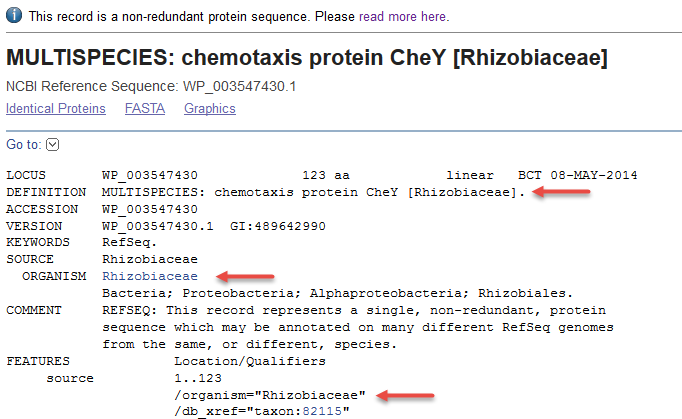
- The lowest common taxonomic node is indicated when a non-redundant protein is found in multiple species (example WP_003547430.1).
- Two organism blocks are provided for proteins that are annotated on genomes from different kingdoms (example WP_041895446.1).
- REFERENCE: this information is not provided. Organism-specific publications can be found on the annotated RefSeq genome records.
- COMMENT: Records include the following standard comment:
- REFSEQ: This record represents a single, non-redundant, protein sequence which may be annotated on many different RefSeq genomes from the same, or different, species.
- Source feature: one or more source features are provided on non-redundant protein records
- A single source feature is provided when the non-redundant protein is found on genomes within a single super-kingdom (e.g., WP_003547430.1 or WP_000091939.1)
- The organism and NCBI tax_id indicated reflect the lowest common level, for the set of genomes that the protein is annotated on.
- The most specific organism information provided is the genus species binomial; strain-specific information is not included on non-redundant protein records.
- The organism information may be at the genus, family or other taxonomic level. This is accompanied by the inclusion of 'MULTISPECIES' prefix in the DEFINITION line.
- Multiple source features are provided when a non-redundant protein is annotated on genomes from different super-kingdoms. This is accompanied by the 'CROSS_KINGDOM' keyword and multiple ORGANISM blocks (e.g.,WP_041895446.1)
Examples:
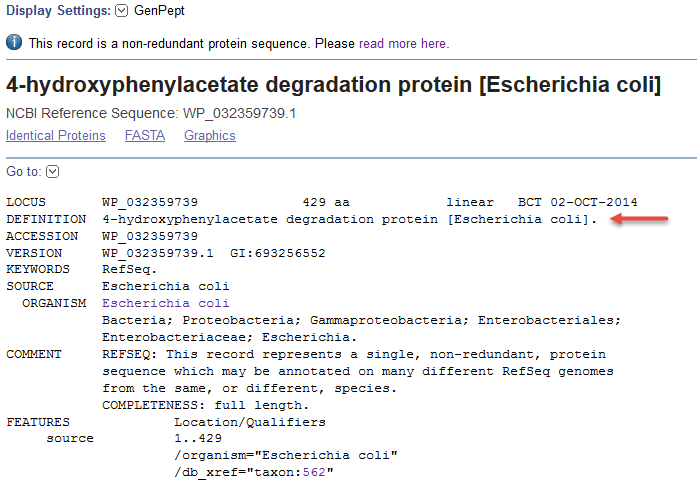
1) Non-redundant RefSeq protein found in a single strain (WP_032359739.1). The DEFINITION, ORGANISM, and source features report the species.
2) Non-redundant RefSeq protein found in multiple species of the genus Rhizobiaceae (WP_003547430.1). The DEFINITION, ORGANISM, and source features report the genus.
3) Non-redundant RefSeq protein found in genomes from both the Archaea and Bacteria super-kingdoms (WP_041895446.1). Note that the DEFINITION line reports two super-kingdoms, each within square brackets. There are two ORGANISM blocks, and there are two source feature blocks.
Links to Related Information
Related Information: Online displays of protein records present a navigation panel on the right side of the web page. The column facilitates access to analysis tools and provides easy access to related resources. Three of the many links that may be provided for non-redundant proteins are indicated with red arrows (shown below).
- Protein clusters: Navigate to NCBI's Protein Clusters resource where you can find information about non-redundant RefSeq proteins that have sequence similarity, as identified by protein clustering methods, but which are not 100% identical. The cluster report for WP_003547430.1 represents a set of 227 similar RefSeq proteins.
- Genomic records: Navigate to the nucleotide database to access, in Summary format, the set of RefSeq genomic sequences that include a CDS feature annotation which encodes the identical non-redundant protein record. Following the "Genomic records" link from WP_003547430.1 to NCBI's Nucleotide resource returns 47 genomic records(as of March 2015). An annotated CDS feature on each of these sequence records cross-references the same non-redundant protein accession and reports the protein name as found on the non-redundant protein record. Should the protein name be updated on WP_003547430.1 then all genomic records that cross-reference this protein will automatically reflect the updated name information. Shown below, is the CDS feature that is annotated on the RefSeq genomic record NC_007761.1.

- Species level organisms: Navigate to NCBI's Taxonomy database to find information on the set of species in which the non-redundant protein has been annotated. As of March 2015, WP_003547430.1 has been annotated on 47 genomes from 16 different species(or 'sp.' organisms).

Identical Protein Report
This report can be accessed using the link provided at the top of the page in the Protein resource. The 'Display Settings' menu also provides access to this report. This display option is available for all protein records available in NCBI's Protein resource.

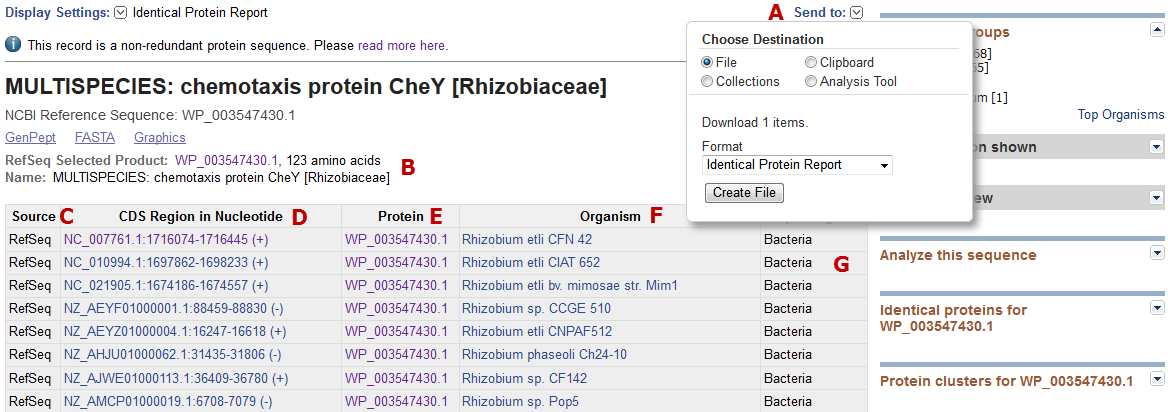
The top section of the Identical protein report for WP_003547430.1 is shown below. The report includes the following information and features:
- A: Report contents can be downloaded as a tabular file for local use.
- B: The top section of the report page shows the protein accession and title of the record being viewed. Beneath that (B) the page shows information for an automatically selected preferred protein accession ('RefSeq Selected Product'), the protein length, and the name as it appears on the DEFINITION line. If you access the Identical Protein report page from a conceptually translated INSDC CDS feature (e.g., AAD28577.1) then you will see that accession number and record title at the top of the Identical protein report and the RefSeq accession and title (B) beneath that.
- C: The database source is indicated. This may be RefSeq, INSDC (conceptual translations from sequences submitted to members of the International Nucleotide Sequence Database collaboration), Swiss-Prot, PDB, PIR, or Patent.
- D, E: The genomic accession and coordinates shown in column 'D' correspond to the annotated CDS feature location which contains a cross-reference to the protein accession shown in 'E'. The link in column D navigates to the Nucleotide database to display the annotated CDS region in GenBank format. The link in column D returns the protein accession indicated in GenPept format.
- F: The Organism column reports information corresponding to the NCBI Taxonomy identifier that is annotated on the Nucleotide record.
- G: The super-kingdom is indicated.